-
PDF
- Split View
-
Views
-
Cite
Cite
Jumana AlHaj Abed, Connie L Cheng, Chase R Crowell, Laura L Madigan, Erica Onwuegbuchu, Siddhi Desai, Judith Benes, Richard S Jones, Mapping Polycomb Response Elements at the Drosophila melanogaster giant Locus, G3 Genes|Genomes|Genetics, Volume 3, Issue 12, 1 December 2013, Pages 2297–2304, https://doi.org/10.1534/g3.113.008896
Close - Share Icon Share
Abstract
Polycomb-group (PcG) proteins are highly conserved epigenetic transcriptional regulators. They are capable of either maintaining the transcriptional silence of target genes through many cell cycles or enabling a dynamic regulation of gene expression in stem cells. In Drosophila melanogaster, recruitment of PcG proteins to targets requires the presence of at least one polycomb response element (PRE). Although the sequence requirements for PREs are not well-defined, the presence of Pho, a PRE-binding PcG protein, is a very good PRE indicator. In this study, we identify two PRE-containing regions at the PcG target gene, giant, one at the promoter, and another approximately 6 kb upstream. PRE-containing fragments, which coincide with localized presence of Pho in chromatin immunoprecipitations, were shown to maintain restricted expression of a lacZ reporter gene in embryos and to cause pairing-sensitive silencing of the mini-white gene in eyes. Our results also reinforce previous observations that although PRE maintenance and pairing-sensitive silencing activities are closely linked, the sequence requirements for these functions are not identical.
Polycomb-group (PcG) genes were initially identified as repressors of Hox genes in Drosophila melanogaster (Lewis 1978). In recent years, the protein products of these epigenetic regulators have been shown to be localized at hundreds of chromosomal sites (Negre et al. 2006; Schwartz et al. 2006; Tolhuis et al. 2006; Oktaba et al. 2008). The majority of their target genes are regulators of development, cell-cycle progression, and/or cell signaling (Oktaba et al. 2008; Classen et al. 2009; Martinez et al. 2009). In PcG mutants, target genes are expressed outside their usual domains of expression, leading to defects in body development. The most conspicuous examples are homeotic transformations that are caused by ectopic expression of Hox genes (Struhl 1983; Wedeen et al. 1986; Simon et al. 1992). Mammalian PcG proteins are required to maintain the pluripotent state of stem cells and their misexpression contributes to a wide variety of human cancers (Richly et al. 2011).
Three major Drosophila PcG protein complexes have been described, Polycomb repressive complex 1 (PRC1), PRC2, and Pleiohomeotic (Pho)-RC. Additional Drosophila complexes that contain PcG proteins dRAF and PR-DUB also have been identified (Lagarou et al. 2008; Scheuermann et al. 2010). Simon and Kingston (2013) recently presented a thorough review of PcG complexes and their activities. Pho-RC consists of the sequence-specific DNA binding protein Pho and Scm-related gene containing four mbt domains (dSfmbt) (Klymenko et al. 2006). The core components of PRC1 are Polycomb (Pc), Polyhomeotic (Ph), Posterior sex combs (Psc), and Sex combs extra (Sce, also known as dRing) (Shao et al. 1999; Saurin et al. 2001). The core components of PRC2 are Enhancer of zeste [E(z)], Extra sex combs (Esc), Suppressor of zeste 12 [Su(z)12], and NURF55 (Czermin et al. 2002; Müller et al. 2002). Multiple variants of Drosophila and mammalian PRC1 and PRC2 complexes have been identified with alternative subunit compositions, which may confer distinct biochemical activities (Simon and Kingston 2013).
Based on examination of the interdependencies of components of Pho-RC, PRC1, and PRC2 for target site binding, a hierarchical binding pathway has been proposed (Wang et al. 2004b). After DNA binding by Pho, PRC2 is recruited by direct interaction with Pho. The E(z) subunit of PRC2 then trimethylates histone H3 at lysine 27 (H3K27me3), facilitating binding by PRC1 because of the affinity of the Pc chromo domain for H3K27me3 (Fischle et al. 2003; Min et al. 2003). However, it is likely that PRC2-independent PRC1 recruitment pathways also exist at some loci (Schwartz et al. 2006). PRC1 may contribute to transcriptional repression by a variety of mechanisms that include monoubiquitylation of histone H2A (H2Aub1), localized chromatin compaction, and/or inhibitory interactions with transcription machinery (Francis et al. 2004; Wang et al. 2004a; Lehmann et al. 2012).
In Drosophila, recruitment of PcG proteins to specific chromosomal sites and repression of nearby genes require the presence of one or more polycomb response elements (PREs) (Simon et al. 1993). Although PcG proteins have been shown to be present at hundreds of genomic locations, PREs at fewer than 20 genes have been functionally characterized. A number of DNA-binding factors have been identified that may contribute to PRE function, including Pho (Brown et al. 1998; Fritsch et al. 1999), Pleiohomeotic-like (Phol) (Brown et al. 2003), GAGA factor (Gaf) (Horard et al. 2000; Mahmoudi et al. 2003), Pipsqueak (Psq) (Lehmann et al. 1998), Zeste (z) (Dejardin and Cavalli 2004), Sp1/Kruppel-like factor (Spps) (Brown et al. 2005; Brown and Kassis 2010), Dorsal switch protein 1 (Dsp1) (Dejardin et al. 2005), and Grainyhead (Grh), (Blastyak et al. 2006), but the exact sequence requirements for PRE activity remain elusive (Kassis and Brown 2013). This is attributable to the heterogeneous sequence organization of PREs and low conservation of consensus sequences for the binding sites of PRE-binding proteins. Among the PRE-binding proteins, only Pho has been detected at all characterized PREs (Kassis and Brown 2013). The other factors appear to be present in various combinations at different PREs. Therefore, Pho localization is a very good indicator of the presence of a biologically functional PRE.
Although the locations of PREs predicted by bioinformatics approaches have shown a low correlation with the distribution of PcG proteins in ChIP-on-chip studies (Ringrose et al. 2003; Fielder and Rehmsmeier 2006; Schwartz et al. 2006; Oktaba et al. 2008), PREs can be functionally defined by their ability to regulate expression of a reporter gene within the context of a transgene. In the PRE maintenance assay, DNA fragments are tested for their abilities to limit enhancer-driven expression of a reporter gene to the normal expression domains of the endogenous gene of the enhancer. This approach has been possible mainly because PREs have been shown to be able to regulate activities of heterologous promoters and enhancers. For example, in the absence of a PRE, enhancers from genes such as Ultrabithorax (Ubx) or engrailed (en) produce ectopic expression of a lacZ reporter, in addition to expression within the normal domains of Ubx and en, respectively. Inclusion of a PRE within the transgene prevents ectopic expression and maintains lacZ expression within the normal boundaries of the endogenous gene (Busturia et al. 1997; Americo et al. 2002; Cunningham et al. 2010; Park et al. 2012).
PREs also are able to repress expression of the mini-white reporter gene, resulting in lighter eye color. Furthermore, the majority of PREs produce a phenomenon known as pairing-sensitive silencing (Kassis et al. 1991). Normally, the eye pigmentation of flies with two copies of a mini-white–containing transgene is approximately twice that of flies with only one copy of the transgene. Pairing-sensitive silencing is observed when flies that are homozygous for the transgene have lighter eye colors compared to heterozygotes. However, not all DNA fragments that function as PREs in maintenance assays also produce pairing-sensitive silencing. For example, the Mcp core PRE requires additional sequences to act as a pairing-sensitive silencer (Muller et al. 1999).
giant (gt) is a zygotic gap gene that affects the development of the head and abdominal regions in Drosophila. Its identification as a PcG target gene was based on the isolation of dominant E(z) alleles that suppress the nanos (nos) maternal effect (Pelegri and Lehmann 1994). The Su(nos) phenotype was shown to be attributable to failure to maintain repression of the gap genes gt and knirps initiated by maternal Hunchback (Hb). ChIP-on-chip studies have confirmed the presence of PcG proteins at gt in embryos (Negre et al. 2006; Oktaba et al. 2008). Initially, gt is expressed in two broad stripes at the syncytial blastoderm stage. By the cellular blastoderm stage, the anterior stripe is divided into two stripes and a patch of expression at the anterior tip is observed (Eldon and Pirrotta 1991). This expression is controlled by four enhancers located upstream of the gt promoter (Berman et al. 2002; Schroeder et al. 2004): gt_(-3) (−1.3 to −2.5 kb) produces the posterior stripe; gt_(-10) (−8.8 to −10.5 kb) produces the major anterior stripes; gt_(-6) (−4.3 to −6.5 kb) produces the anterior tip expression; and gt_(-1) (−0.05 to −1.3 kb) produces both the posterior and major anterior stripes. The role played by the apparent redundancy of gt_(-1) with gt_(-10) and gt_(-3) in regulation of the endogenous gt gene is not clear. Beyond gastrulation, gt primarily is expressed in the head region (Eldon and Pirrotta 1991). Through the cellular blastoderm stage, it appears that PcG repression of gt is redundant with repression by gene-specific transcription factors, such as Hb (Pelegri and Lehmann 1994). However, Pc mutants exhibit ectopic gt expression in later embryonic stages (Negre et al. 2006). To initiate a more detailed analysis of PcG regulation of gt, we have mapped the locations of gt PREs.
Materials and Methods
Drosophila stocks and generation of transgenic lines
Strains are described at the Bloomington Drosophila Stock Center web site (http://flystocks.bio.indiana.edu) unless otherwise specified. To produce the SD10 constructs, gt genomic fragments were PCR-amplified from CH322-101F2 or CH322-5H16 BAC clones (BACPAC resources: http://www.pacmanfly.org/) using primers that included FRT sequences and NspI sites (Supporting Information, Table S1). Amplified fragments were digested with NspI and ligated to the vector SphI site located between the en enhancer and promoter (in the same orientation relative to the en promoter as in the gt promoter). SD10-gt constructs were injected into w1118 embryos by Genetics Services (Sudbury, MA) or BestGene (Chino Hills, CA). Additional lines were obtained by transposon mobilization using P[Δ2-3] (Robertson et al. 1988). The gt fragments were deleted from transgenes by crossing SD10-gt females to hsFLP males and heat-shocking embryos or first instar larvae for 1 hr at 37°. Deletions were verified by PCR using SD10C-U (5′GTTGAGCCGAAGAGAAAATACGC-3′) and SD10CL (5′-GTTTTCCCACTCACGACGTTG-3′) primers. To examine β-galactosidase expression in a PcG-mutant background, SD10-gt males were crossed to ph-d401 ph-p602 w1/FM7c females. To test for pairing-sensitive silencing, males were aged 2 days after eclosure and the eye colors of flies that were homozygous for the transgene were compared to those of flies that were heterozygous for the same transgene.
Immunostaining of embryos
Embryos were collected at 25° then were fixed and processed essentially as previously described (Jones and Gelbart 1990) with the following modifications. Rabbit anti-β-galactosidase antibodies (Cappel) were diluted 1:1500. Biotin-SP–conjugated goat-anti-rabbit secondary antibodies (Jackson Immunoresearch) were diluted 1:10,000. Streptavidin-horseradish peroxidase (Jackson Immunoresearch) was diluted 1:5000. Signals were detected by incubating embryos in 1 mg/ml diaminobenzidine (Sigma-Aldrich) in 0.1 M Tris-HCl (pH 7), 1% NiCl2, and 0.003% H2O2 for approximately 20 min. Reactions were stopped by washing with PBST (0.01% Trition-X-100). Embryos were dehydrated and mounted in Permount (Fisher Scientific). Images were obtained using a Ziess Axiovert 200M microscope.
Chromatin immunoprecipitation
The protocol was performed as indicated in the ChIP assay kit (Millipore) with the following modifications. Oregon-R embryos were collected for 30 min, aged for 170 min at 25°, and then dechorionated in 50% hypochlorite bleach, briefly washed with water, and fixed in 2% formaldehyde in PBS:heptane (1:3) for 20 min. Fixation was quenched by addition of glycine to a final concentration of 50 mM. Embryos were then washed, weighed, flash-frozen, and stored at −80°. Embryos were homogenized in 50 mM Tris (pH 8.1) and 10 mM EDTA supplemented with 1.25× complete EDTA-free protease inhibitor cocktail (Roche). SDS was added to a final concentration of 1%. After 10 min of incubation on ice, samples were sonicated in a cup horn sonicator (Misonix sonicator 3000) at 4° to produce DNA fragments that were predominantly within the range of 0.3 to 0.7 kb. An equivalent of 2 mg of embryos was set aside as input genomic DNA and incubated with 0.2 M NaCl overnight at 65° to reverse the crosslinks. The remainder of the supernatant was diluted 10-fold with ChIP dilution buffer, and chromatin from the equivalent of 3.2 mg of embryos was incubated with 2.5 μl rabbit anti-Pho antisera (Brown et al. 2003) or 5 μl rabbit pre-immune serum for mock.
Quantitative PCR
Quantitative PCR was performed with PerfeCTa SYBR Green SuperMix (Quanta Biosciences) using a Rotor Gene RG3000 thermocycler (Corbett Research). Immunoprecipitated DNA from 100 μg of embryos was used for each PCR reaction. Sequences of primers are in Table S2. PCR reactions were performed in triplicate for each ChIP experiment. The data presented are the average of three independent ChIP experiments from independent chromatin preparations. Percent input values were calculated using Rotor Gene software by comparing Ct values of samples to standard curves established by the Ct values of total chromatin controls.
Eye pigment assay
Eye pigmentation was quantified by homogenizing a total of 10 male heads (4 days after eclosion) from each fly group in 0.5 ml of 0.01 M HCl in ethanol. Homogenate was left overnight at 4°, warmed for 5 min at 50°, and then centrifuged. The absorbance of the supernatant was measured at 480 nm (Pal-Bhadra et al. 2004). Assays for each genotype were performed in triplicate. P values were calculated using unpaired t test to determine statistical significance.
Results
Pho binds to two locations within gt cis-regulatory region
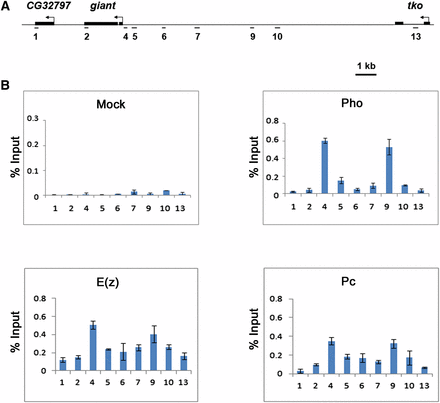
To date, all PREs that have been functionally tested bind Pho (Oktaba et al. 2008; Kassis and Brown 2013). Therefore, the presence of Pho at a given genomic region is a good indicator of the location of a biologically active PRE. To identify gt PREs, we began by precisely mapping the distribution of Pho across a 19-kb region that encompasses the gt locus and extends into flanking genes CG32797 and technical knockout (tko) (Figure 1A). ChIP assays were performed on Oregon-R blastoderm stage embryos using anti-Pho antibodies. Pho was detected at two regions within the gt upstream regulatory region of gt (Figure 1B). The first is near the promoter at the PCR-amplified region 4 (+9 to −168). The second is approximately 6 kb upstream at region 9 (−6108 to −6320) (Figure 1). These results are consistent with a genome-wide ChIP-on-chip study that reported the presence of Pho at the gt locus in embryos (Oktaba et al. 2008). ChIP assays with anti-E(z) and anti-Pc antibodies show colocalization of these PRC2 and PRC1 subunits with Pho, but with broader distributions (Figure 1B). Even though the presence of Pho at these regions is highly suggestive that they contain PREs, in vivo tests, such as PRE maintenance or pairing-sensitive silencing assays, are required to confirm these predictions.
Pho binds to two regions within the giant cis-regulatory region that colocalizes with peaks of E(z) and Pc distribution. (A) Schematic representation showing the giant genomic region and flanking genes. Regions amplified by PCR in ChIP assays (1–13) are shown. Region 1 and 13 are, respectively, within the CG32797 and tko genes and serve as negative controls. (B) ChIP analysis of Oregon-R embryos 2 h 50 min to 3 h 20 min after egg lay shows two Pho peaks, one close to the gt promoter (region 4) and the other ∼6 kb upstream (region 9). E(z) and Pc, subunits of PRC2 and PRC1, respectively, colocalize with Pho but are more broadly distributed. Preliminary ChIP assays using 33 primer sets spanning this 19-kb region did not show the presence of Pho at additional sites (data not shown). ChIP was performed as indicated with anti-Pho antibody (upper right panel), anti-E(z) antibody (lower left panel), anti-Pc antibody (lower right panel), and rabbit preimmune antiserum for mock (upper left panel). ChIP signals are presented as a percent of input chromatin. Error bars represent SD.
gt has two PRE-containing regions that coincide with Pho localization
To test for PRE activity, gt genomic fragments were inserted into the SD10 P-element vector, which contains an engrailed (en) enhancer and promoter upstream of the lacZ reporter gene. The en enhancer produces lacZ expression in 14 stripes and resembles endogenous en expression. Without the presence of a PRE, ectopic expression of lacZ is detected between en-like stripes. However, the addition of an en PRE (Devido et al. 2008) or a heterologous PRE from the invected (inv) or Ubx (bxdd) genes (Cunningham et al. 2010) results in maintenance of the restricted en-like lacZ expression.
The gt fragments were inserted between the en upstream regulatory region and en promoter to produce SD10-gt constructs (Figure 2B). Each insert was flanked by FRT sites to allow precise excision of gt insert by FLP recombinase in vivo. By comparing reporter expression from the intact transgenes and insert-deleted derivatives, it is possible to distinguish regulatory effects of the gt insert from potential position effect of the genomic location of the transgene.
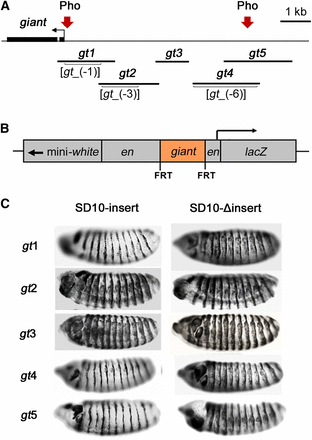
The giant inserts tested for their abilities to maintain en-like expression pattern of β-galactosidase. (A) A schematic of gt upstream regulatory region showing fragments gt1–gt5 that were cloned into the SD10 vector and tested for PRE activity The locations of previously mapped gt enhancers gt_(-1), gt_(-3), and gt_(-6) are indicated in brackets. Pho-positive regions are indicated by arrows and correspond to PCR amplified regions 4 and 9 (Figure 1). (B) A schematic representation of the en-lacZ reporter construct SD10. Inserts are flanked by FRT sites. (C) Stage 14 embryos from transgenic lines stained for β-galactosidase expression. Lateral views of embryos are shown, anterior to the left, dorsal up. Transgenic lines tested are indicated to the left of the embryos. Expression patterns are representative of those produced by multiple lines of each construct. However, lines that failed to maintain the en-like pattern exhibited varying degrees of ectopic expression. Embryos containing intact transgenes are on the left. ΔInsert lines (right) are FLP recombinase–mediated deletion derivatives of the same lines shown on the left.
Guided by the locations of Pho-positive ChIP regions and the locations of gt embryonic enhancers (Berman et al. 2002; Schroeder et al. 2004), five overlapping fragments were selected for analysis (Figure 2A): gt1 includes the first Pho-positive region by ChIP, and the entire gt_(-1) enhancer; gt2 contains the entire gt_(-3) enhancer; gt3 does not include an identified enhancer and does not show Pho binding by ChIP; gt4 contains the gt_(-6) enhancer; and the gt4 and gt5 fragments overlap by 1.2 kb and contain the second Pho-positive region. The constructs were used to produce multiple transgenic lines. The inserts were deleted from a subset of these lines by FLP recombinase. Stage 14 embryos containing intact transgenes and their deletion derivatives were tested for β-galactosidase expression.
Embryos from each of the assayed transgenic lines containing SD10-gt1, SD10-gt4, or SD10-gt5 showed restricted en-like patterns of β-galactosidase expression, indicative of the presence of a PRE within those constructs, whereas all SD10-gt2 and SD10-gt3 lines showed distinct ectopic expression of the lacZ reporter. Representative embryos for each construct are shown for each in Figure 2C. Maintenance of the en-like expression pattern was lost upon excision of gt1, gt4, and gt5 inserts (Figure 2C). Ectopic β-galactosidase expression was observed before and after the deletion of gt2 or gt3 inserts. A summary of all lines tested is shown in Table 1. Based on the results of these PRE maintenance assays, we conclude that the gt1, gt4, and gt5 fragments contain PREs. These results are consistent with the localization of Pho at region 4 within the gt1 fragment and at region 9 within the region of overlap between gt4 and gt5.
Summary of PRE maintenance assay results
| Construct . | Pho . | PRE Function . |
|---|---|---|
| gt1 | + | 5/5 |
| gt2 | − | 0/4 |
| gt3 | − | 0/5 |
| gt4 | + | 5/5 |
| gt5 | + | 2/2 |
| Construct . | Pho . | PRE Function . |
|---|---|---|
| gt1 | + | 5/5 |
| gt2 | − | 0/4 |
| gt3 | − | 0/5 |
| gt4 | + | 5/5 |
| gt5 | + | 2/2 |
A list of SD10-gt constructs indicating whether Pho was found to bind to the respective gt fragments in ChIP assays and the number of lines exhibiting PRE activity in maintenance assays (PRE function).
| Construct . | Pho . | PRE Function . |
|---|---|---|
| gt1 | + | 5/5 |
| gt2 | − | 0/4 |
| gt3 | − | 0/5 |
| gt4 | + | 5/5 |
| gt5 | + | 2/2 |
| Construct . | Pho . | PRE Function . |
|---|---|---|
| gt1 | + | 5/5 |
| gt2 | − | 0/4 |
| gt3 | − | 0/5 |
| gt4 | + | 5/5 |
| gt5 | + | 2/2 |
A list of SD10-gt constructs indicating whether Pho was found to bind to the respective gt fragments in ChIP assays and the number of lines exhibiting PRE activity in maintenance assays (PRE function).
Maintenance of repression by gt fragments is PcG-dependent
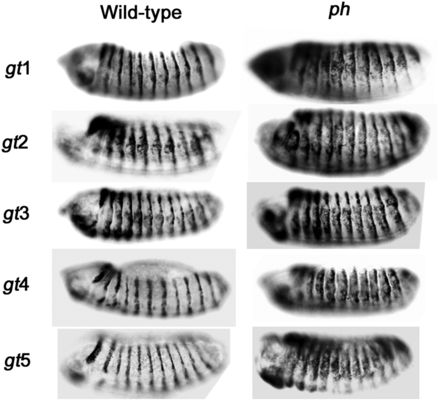
To confirm that the repressive ability of gt fragments is PcG-dependent, β-galactosidase expression was analyzed in a PcG mutant background. Representative transgenic lines for each construct were crossed to a polyhomeotic (ph) mutant (ph-d401 ph-p602 double mutants) and β-galactosidase expression was compared to the same transgenic lines in a wild-type PcG background (Figure 3). SD10-gt1, SD10-gt4, and SD10-gt5 all showed ectopic β-galactosidase expression in the ph mutant. This PcG-dependent maintenance of reporter gene repression confirms that SD10-gt1, SD10-gt4, and SD10-gt5 contain PREs. SD10-gt2 and SD10-gt3 constructs showed ectopic β-galactosidase expression in both ph+ and ph− backgrounds, consistent with lack of a functional PRE in these fragments.
Maintenance of SD10-gt reporter repression is PcG-dependent. Stage 14 embryos from transgenic lines stained for β-galactosidase expression. Embryos with a wild-type PcG background are on the left. Embryos from crosses of transgenic lines to ph-d401 ph-p602 double mutants are on the right. Orientation and identification of embryos are the same as in Figure 2C.
The gt fragments are able to demonstrate pairing-sensitive silencing
Because some, but not all, of the previously tested PREs exhibit pairing-sensitive silencing (Kassis 2002), all SD10-gt transgenes that were able to homozygose were also tested for their ability to demonstrate pairing-sensitive silencing of the mini-white gene in the SD10 vector (Figure 4). The eye colors of transgenic flies homozygous for the transgene were compared to those of the heterozygotes. If a transgene exhibits pairing-sensitive silencing, then homozygotes will show stronger repression of the mini-white gene within the SD10 vector, and as a result will have lighter eye color when compared to heterozygous flies with only one copy of the transgene. Consistent with the presence or lack of PREs based on the PRE maintenance assay (Figure 2C), three out of four SD10-gt1 and four out of five SD10-gt5 lines demonstrated pairing-sensitive silencing compared to zero out of three SD10-gt2 and zero out of five SD10-gt3 lines (Figure 4A). Curiously, although the gt4 fragment overlaps with gt5 and behaves as a PRE in the maintenance assay, only one out of nine SD10-gt4 lines exhibited pairing-sensitive silencing (Figure 4A). Of the transgenic lines for which we have both intact transgenes and their FLP recombinase-induced derivatives (Figure 4A, right column), the results were consistent. Of this subset of lines, two out of two SD10-gt1 lines and two out of two SD10-gt5 lines lost pairing-sensitive silencing on deletion of the gt fragment. The eyes of flies from representative lines are shown in Figure 2, B and C. Deletion of the gt fragment from the sole SD10-gt4 line that produced pairing-sensitive silencing also eliminated this activity (data not shown). This suggests that the region shared by gt4 and gt5 is necessary, but not sufficient, for pairing-sensitive silencing, and that additional sequences contained within gt5, but not gt4, are needed. Quantitative assays performed with the same fly lines shown in Figure 4, B and C showed eye pigment levels that were consistent with the visual appearance of eye colors (Figure 4D).
The gt fragments exhibit pairing-sensitive silencing. (A) The number of transgenic lines exhibiting pairing-sensitive silencing (PSS) relative to total number of lines tested. The middle column lists the results for all lines tested for each construct. The right column lists the results for the subset of lines for which deletion-derivatives were generated. These are the subset of lines that were tested for β-galactosidase expression (Table 1) and that were homozygous-viable. (B) Eye pigmentation of SD10-gt transgenic flies. Specific transgenic lines are indicated to the left. Flies were either heterozygous (P[w+]/+) or homozygous (P[w+]/P[w+]) for the transgene. SD10-gt1 and SD10-gt5 homozygotes showed reduced eye pigmentation compared to heterozygotes (PSS). None of the SD10-gt2 or SD10-gt3 lines exhibited pairing-sensitive silencing. Eight of nine SD10-gt4 lines did not exhibit PSS. (C) Deletion lines corresponding to the same lines in (B) after excision of the gt fragment by FLP recombinase. PSS seen in the SD10-gt1.2 and SD10-gt5.4 homozygotes was lost upon deletion of the gt fragment. (D) Quantitative assays of fly eye pigments of the same lines shown in (B) and (C). yw is the y w67c23 stock that contains the transgenes. The absorbance values for heterozygotes and homozygotes of each transgene are, respectively, illustrated as orange and blue bars. Error bars represent SD. There was a statistically significant difference between absorbance values for heterozygotes and homozygotes of each line. gt4.1 and Δgt4.1, P < 0.05. All other lines, P < 0.001.
Discussion
We have identified and mapped the locations of two PRE-containing regions at the gt locus. One is near the promoter and a second is ∼6 kb upstream. This organization is most similar to the PRE distribution at the invected (inv) gene, which has two PREs located at similar positions relative to the inv transcription start site (Cunningham et al. 2010). inv is part of a gene complex that also includes the engrailed (en) gene. en has two closely linked PREs that are located in an interval spanning ∼0.4 to 2.4 kb upstream of the transcription start site (Americo et al. 2002; Devido et al. 2008). The even skipped (eve) gene also has two PREs; one is near the promoter and the second PRE is ∼9 kb downstream (Fujioka et al. 2008). It has been suggested that multiple PREs at inv, en, and eve may contribute to physical interactions between remote cis-regulatory regions and promoters (Devido et al. 2008; Fujioka et al. 2008; Cunningham et al. 2010). It is likely that this activity may be responsible for the pairing-sensitive silencing observed in the context of transgenes. It is not clear to what extent multiple PREs may be functionally redundant at their endogenous loci. Each is defined by its independent ability to maintain transcriptional repression of a transgenic reporter. However, the observation that more robust repression is produced by a combination of both en PREs than by either PRE alone suggests that they are only partially redundant (Devido et al. 2008). The degree to which gt PREs may be functionally redundant remains to be determined. It is also possible that gt PREs may independently regulate distinct enhancers or may somehow cooperate in the establishment and/or maintenance of PcG-mediated repression.
Even though most PREs that test positive in maintenance assays also show pairing-sensitive silencing, it has been shown that pairing-sensitive silencing does not always correlate with PRE function. In some cases, a pairing-silencing element, conferring pairing-sensitive silencing, is distinct from a PRE. One example is the Mcp PRE, which regulates the Abdominal-B gene of the Bithorax complex. The 800-bp Mcp core fragment behaves as a functional PRE in reporter maintenance assays but is unable to demonstrate pairing-sensitive silencing unless supplemented with additional flanking sequences or possibly other regulatory sequences from other genes (Busturia et al. 1997; Muller et al. 1999). The sequence shared by gt4 and gt5 may be similar to the Mcp core fragment in that both SD10-gt4 and SD10-gt5 maintain repression of en enhancer-driven reporter expression, but only SD10-gt5 exhibits pairing-sensitive silencing. It may be that sequences contained in gt5, but not gt4, include binding sites for proteins needed for pairing-sensitive silencing, but not for maintenance assays. Definition of the sequence requirements for pairing-sensitive silencing will require further dissection of this portion of the gt regulatory region and mutational analysis of potential transcription factor binding sites.
Although attempts have been made to identify PREs based on their sequences (Ringrose et al. 2003; Fielder and Rehmsmeier 2006), recent studies have shown that there are shortcomings to using consensus binding sites to predict the location of PREs (Schwartz et al. 2006; Oktaba et al. 2008; Cunningham et al. 2010). This is partially attributable to the sequence heterogeneity of characterized PREs, which reflects the variable assortment of proteins that bind to any given PRE. With the exception of Pho, only a subset of identified PRE binding proteins appears to bind to any particular PRE. Furthermore, the binding sites for several of these proteins are highly variable. For example, Pho consensus binding sites do not always correlate to Pho binding or PRE function. The core of the Pho consensus sequence was originally defined as GCCAT (Fritsch et al. 1999). On the basis of genome-wide ChIP-on-chip studies, an expanded consensus sequence has been proposed, G(C/A)(C/G)GCCAT(T/C)TT (Oktaba et al. 2008). Even more perplexing, Pho has been shown to bind to fragments that contain neither of these consensus sequences (Cunningham et al. 2010).
Examination of the gt1, gt4, and gt5 sequences reveals that, although Pho consensus sites coincide with the ChIP-defined localization of Pho in embryos (Figure 1B), Pho does not appear to bind to regions that contain many additional Pho consensus sites (Figure 5 and Table 2). For example, the gt2 and gt3 fragments have eight and seven Pho consensus sites, respectively, but are negative for Pho binding in ChIP assays and fail to behave as PREs in the maintenance and pairing-sensitive silencing assays. The extended Pho-consensus sequence (Oktaba et al. 2008) is not present within the gt regulatory region. It seems probable that a PRE is more likely to be determined by the presence of multiple binding sites for an array of DNA-binding proteins, and that Pho binding may involve cooperative interactions with some combination of other DNA binding factors (Kassis and Brown 2013). It is also possible that Pho may be bound to other regions in the gt cis-regulatory region at later developmental stages.
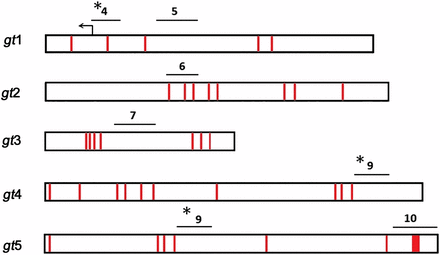
Locations of Pho consensus binding sites within gt fragments. Schematic of gt fragments with Pho core consensus sites (GCCAT) in red. Lines above each gt fragment represent regions amplified by PCR in ChIP assays. Pho-positive regions (see Figure 1B) are indicated with asterisks.
Summary of Pho consensus sites within gt fragments
| Fragment . | Coordinates . | Pho Consensus Sites . |
|---|---|---|
| gt1 | +274 to −1629 | 5 |
| gt2 | −1143 to −3152 | 8 |
| gt1, 2 overlap | −1143 to −1629 | 0 |
| gt3 | −3032 to −4150 | 7 |
| gt4 | −4302 to −6539 | 10 |
| gt5 | −5341 to −7641 | 10 |
| gt4, 5 overlap | −5341 to −6539 | 4 |
| Fragment . | Coordinates . | Pho Consensus Sites . |
|---|---|---|
| gt1 | +274 to −1629 | 5 |
| gt2 | −1143 to −3152 | 8 |
| gt1, 2 overlap | −1143 to −1629 | 0 |
| gt3 | −3032 to −4150 | 7 |
| gt4 | −4302 to −6539 | 10 |
| gt5 | −5341 to −7641 | 10 |
| gt4, 5 overlap | −5341 to −6539 | 4 |
Summary of Pho consensus sites within gt fragments. Coordinates of gt fragments relative to the transcription start site and summary of Pho consensus sites within each fragment. gt1 and gt2 overlap by 486 bp. gt4 and gt5 overlap by 1198 bp.
| Fragment . | Coordinates . | Pho Consensus Sites . |
|---|---|---|
| gt1 | +274 to −1629 | 5 |
| gt2 | −1143 to −3152 | 8 |
| gt1, 2 overlap | −1143 to −1629 | 0 |
| gt3 | −3032 to −4150 | 7 |
| gt4 | −4302 to −6539 | 10 |
| gt5 | −5341 to −7641 | 10 |
| gt4, 5 overlap | −5341 to −6539 | 4 |
| Fragment . | Coordinates . | Pho Consensus Sites . |
|---|---|---|
| gt1 | +274 to −1629 | 5 |
| gt2 | −1143 to −3152 | 8 |
| gt1, 2 overlap | −1143 to −1629 | 0 |
| gt3 | −3032 to −4150 | 7 |
| gt4 | −4302 to −6539 | 10 |
| gt5 | −5341 to −7641 | 10 |
| gt4, 5 overlap | −5341 to −6539 | 4 |
Summary of Pho consensus sites within gt fragments. Coordinates of gt fragments relative to the transcription start site and summary of Pho consensus sites within each fragment. gt1 and gt2 overlap by 486 bp. gt4 and gt5 overlap by 1198 bp.
In conclusion, we have demonstrated the presence of two gt PRE-containing regions within the gt cis-regulatory region. Consistent with other characterized PREs, both gt PRE-containing regions correspond to localized binding by Pho in vivo. The presence of Pho at a single location within each of these regions indicates that it is likely that each contains just one PRE. However, at this time we cannot rule out the possibility that one or both regions may contain multiple closely linked PREs.
Acknowledgments
We thank Judy Kassis for providing the SD10 vector, the hsFLP fly stock, and helpful discussions. We also thank Basilia Oseguero and Alexandria McIlveene for assistance with fly experiments and maintenance of fly stocks. The ph-d401 ph-p602 fly stock was obtained from the Bloomington stock center. This work was supported by National Institutes of Health grant R15GM094737 (to R.S.J.).
Footnotes
Communicating editor: J. A. Birchler
Literature Cited
Author notes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.113.008896/-/DC1



![The gt fragments exhibit pairing-sensitive silencing. (A) The number of transgenic lines exhibiting pairing-sensitive silencing (PSS) relative to total number of lines tested. The middle column lists the results for all lines tested for each construct. The right column lists the results for the subset of lines for which deletion-derivatives were generated. These are the subset of lines that were tested for β-galactosidase expression (Table 1) and that were homozygous-viable. (B) Eye pigmentation of SD10-gt transgenic flies. Specific transgenic lines are indicated to the left. Flies were either heterozygous (P[w+]/+) or homozygous (P[w+]/P[w+]) for the transgene. SD10-gt1 and SD10-gt5 homozygotes showed reduced eye pigmentation compared to heterozygotes (PSS). None of the SD10-gt2 or SD10-gt3 lines exhibited pairing-sensitive silencing. Eight of nine SD10-gt4 lines did not exhibit PSS. (C) Deletion lines corresponding to the same lines in (B) after excision of the gt fragment by FLP recombinase. PSS seen in the SD10-gt1.2 and SD10-gt5.4 homozygotes was lost upon deletion of the gt fragment. (D) Quantitative assays of fly eye pigments of the same lines shown in (B) and (C). yw is the y w67c23 stock that contains the transgenes. The absorbance values for heterozygotes and homozygotes of each transgene are, respectively, illustrated as orange and blue bars. Error bars represent SD. There was a statistically significant difference between absorbance values for heterozygotes and homozygotes of each line. gt4.1 and Δgt4.1, P < 0.05. All other lines, P < 0.001.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/g3journal/3/12/10.1534_g3.113.008896/5/m_2297f4.gif?Expires=1716323949&Signature=TIQeRxtnAR9Hw1~A8pluCX-AhANuwJc0Hf~j2s0nXDVSPWnXuUarkdSA85yeSYUCZvkfewtPgLK1sX4vuCkSPdFu3RNl0b48mb-e5rZpjpaC6JXlU9pxQMJ-9KVoofjTXXXzyfvtgpT5E1w-Ht97MMBllZqinHYha9KmpOwluwC4Nnr6GuSUCsN7btPzAQpKuMQclR9DbfUvfta2AmWUIaZegfxU5lsY6-sSnG2hhNFvbxyDmh~N2xWUbEXUBAOEN5r1BCNakGvNcLNKox~SRfvFSV0faW5v25kIGnS9hDWg6iihrV2LMfbJcO5gl0UY5mSYvQs27ULnen~4Qnbkgw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)