-
PDF
- Split View
-
Views
-
Cite
Cite
Sangmi Oh, Ichiro Kawasaki, Jae-Hyung Park, Yhong-Hee Shim, cdc-25.4, a Caenorhabditis elegans Ortholog of cdc25, Is Required for Male Mating Behavior, G3 Genes|Genomes|Genetics, Volume 6, Issue 12, 1 December 2016, Pages 4127–4138, https://doi.org/10.1534/g3.116.036129
Close - Share Icon Share
Abstract
Cell division cycle 25 (cdc25) is an evolutionarily conserved phosphatase that promotes cell cycle progression. Among the four cdc25 orthologs in Caenorhabditis elegans, we found that cdc-25.4 mutant males failed to produce outcrossed progeny. This was not caused by defects in sperm development, but by defects in male mating behavior. The cdc-25.4 mutant males showed various defects during male mating, including contact response, backing, turning, and vulva location. Aberrant turning behavior was the most prominent defect in the cdc-25.4 mutant males. We also found that cdc-25.4 is expressed in many neuronal cells throughout development. The turning defect in cdc-25.4 mutant males was recovered by cdc-25.4 transgenic expression in neuronal cells, suggesting that cdc-25.4 functions in neurons for male mating. However, the neuronal morphology of cdc-25.4 mutant males appeared to be normal, as examined with several neuronal markers. Also, RNAi depletion of wee-1.3, a C. elegans ortholog of Wee1/Myt1 kinase, failed to suppress the mating defects of cdc-25.4 mutant males. These findings suggest that, for successful male mating, cdc-25.4 does not target cell cycles that are required for neuronal differentiation and development. Rather, cdc-25.4 likely regulates noncanonical substrates in neuronal cells.
Cdc25 phosphatase promotes cell cycle progression by removing an inhibitory phosphate from cyclin-dependent kinase 1 (Cdk1), which is placed by Wee1/Myt1 kinase (Donzelli and Draetta 2003) . Cdc25 is a highly conserved phosphatase that is present from yeast to mammals (Edgar and O’Farrell 1989; Jimenez et al. 1990; Sadhu et al. 1990; Forsburg and Nurse 1991). In C. elegans, four cdc-25 genes, cdc-25.1, cdc-25.2, cdc-25.3, and cdc-25.4, have been identified (Ashcroft et al. 1998). cdc-25.1 and cdc-25.2 are required for germline mitotic proliferation (Ashcroft et al. 1999; Ashcroft and Golden 2002; Kim et al. 2009; Yoon et al. 2012) and for oocyte meiotic maturation (Kim et al. 2010), respectively. The roles of cdc-25 genes are not confined to the germ cells. cdc-25.1 acts at the G1/S transition in the embryonic E lineage, and stabilization of CDC-25.1 in the E lineage in cdc-25.1(gf) mutants causes an increase in the number of intestinal cells (Hebeisen and Roy 2008; Segref et al. 2010). cdc-25.2 is also required for proper progression of characteristic intestinal divisions (Lee et al. 2016). These results demonstrate that cdc-25 genes are involved in both germline and somatic development. However, the functions of cdc-25.4 remain largely unknown.
Intriguingly, the functions of these cell cycle regulators are not limited to control of the cell cycle. A variety of cell cycle regulators are now known to be involved in other biological processes, including regulation of transcription, epigenetics, metabolism, and neuronal functions (Lim and Kaldis 2013). For example, CDK10 suppresses the transcriptional activity of the transcription factor ETS2 via a direct interaction in human cells (Kasten and Giordano 2001). CDK1 and CDK2 phosphorylate enhancer of zeste homolog 2, the catalytic subunit of Polycomb repressive complex 2, in human prostate cancer cells, and this phosphorylation regulates the methyltransferase activity of the complex, which leads to transcriptional repression of target genes (Chen et al. 2010). The functions of CDK5 in neuronal development in multiple organisms have been reported (Su and Tsai 2011). In C. elegans, CDK-5 regulates polarized trafficking of presynaptic components and dense-core vesicles (Ou et al. 2010; Goodwin et al. 2012). CDK-5 is also required for synapse elimination and formation (Park et al. 2011), and for trafficking of glutamate receptors in the ventral nerve cord (Monteiro et al. 2012). CDK-8 is required for axon navigation decisions in neurons (Steimel et al. 2013), and also functions in vulval development by regulating the epidermal growth factor receptor-Ras-extracellular signal-regulated kinase pathway (Grants et al. 2016). Taken together, these results suggest that CDKs phosphorylate not only cell cycle regulators, but also other regulators that are involved in diverse, important biological processes.
Male mating behavior, which is regulated by multiple neuronal networks, is one of the most complex behaviors observed in C. elegans. C. elegans contains a limited number of neuronal cells. Hermaphrodites have 302 neurons out of 959 somatic cells, and males have 385 neurons out of 1031 somatic cells (Sulston and Horvitz 1977). Among them, 170 neurons, which include 81 male-specific neurons and 89 neurons common to both sexes, function in male mating processes, in collaboration with 64 muscles (Jarrell et al. 2012). Notably, most of these cells are located in the male tail region (Sulston et al. 1980). The male mating process can be divided into several stages as follows: (1) recognition of a hermaphrodite by touching her body with his tail, (2) moving backward until reaching either the head or tail of her body, (3) turning, (4) location of his tail at the vulva, (5) spicule insertion, and (6) ejaculation (Loer and Kenyon 1993; Liu and Sternberg 1995). All of these steps are closely associated with each other and integrated together in the nervous system via male-specific neurons including rays, CP, hook, and postcloacal sensilla neurons (Barr and Garcia 2006).
Here, we report, for the first time, that cdc-25.4 regulates male mating behavior. We found that cdc-25.4 is expressed in many neurons throughout development. Depletion of cdc-25.4 activity resulted in male sterility caused by defects in male mating behavior. The turning defect was the most prominent defective phenotype displayed by cdc-25.4 mutant males. This defect was rescued by cdc-25.4 transgene expression driven by the endogenous cdc-25.4 promoter, as well as by several neuronal promoters, indicating that cdc-25.4 activity is required in neurons for successful male mating. Furthermore, cdc-25.4 mutant males showed several additional defects, such as contact response, backing, and vulva location defects, suggesting that cdc-25.4 is required in multiple processes during mating. Interestingly, the turning defect of cdc-25.4 mutant males was not suppressed by depletion of wee-1.3, unlike canonical cell cycle regulations by CDC-25.1 and CDC-25.2, such as those in germline mitotic proliferation, oocyte maturation, and intestinal divisions, in which defects of cdc-25 mutants were largely suppressed by depletion of wee-1.3, a C. elegans ortholog of Wee1/Myt1 kinase (Kim et al. 2010; Yoon et al. 2012; Lee et al. 2016). This result suggests that cdc-25.4 does not function in the regulation of the cell cycle in the male nervous system. Rather, cdc-25.4 may have a unique noncanonical function in neuronal cells that is vital for successful male mating behavior.
Materials and Methods
Strains and maintenance
C. elegans strains were grown as described previously (Brenner 1974). The Bristol N2 strain or him-5(e1467) was used as wild-type controls for all experiments. The mutant alleles used in this study were as follows: LGI; ttTi4348; LGII; cdc-25.4(tm4088), cat-2(e1112); LGIII; glp-1(q231), unc-119(ed3), bas-1(ad446); LGIV; fem-1(hc17), fem-3(q20), unc-22(e66); and LGV; fog-2(q71), him-5(e1467). The following transgenic reporters were used in this study: egIs1[dat-1p::GFP], kkuIs05[Pcdc-25.4::gfp::cdc-25.4::cdc-25.4 3′ UTR; cb-unc-119(+)] I, kkuEx06[Pcdc-25.4::gfp::cdc-25.4 3′ UTR; Prab-3::mCherry::unc-54 3′ UTR; cb-unc-119(+)], kkuEx08[Ptph-1::gfp::cdc-25.4::unc-54 3′ UTR; Pmyo-2::mCherry::unc-54 3′ UTR], kkuEx09[Punc-119::gfp::cdc-25.4::unc-54 3′ UTR; Pmyo-2::mCherry::unc-54 3′ UTR], kkuEx15[Ptba-9::gfp::cdc-25.4::unc-54 3′ UTR; Pmyo-2::mCherry::unc-54 3′ UTR], kkuEx16[Ppkd-2::gfp::cdc-25.4::unc-54 3′ UTR; Pmyo-2::mCherry::unc-54 3′ UTR], mgIs42[tph-1p::GFP + pRF4(rol-6(su1006))], otIs45[unc-119::GFP] V, oxEx1580[eft-3p::GFP; cb-unc-119(+)], and uIs60[unc-119p::YFP + unc-119p::sid-1]. All strains were maintained at either 15 or 20° on nematode growth medium (NGM) agar plates containing Escherichia coli strain OP50. Males were generated either by introducing the him-5(e1467) mutation to the strain or by heat shock (Hodgkin 1983).
Quantitative real-time RT-PCR
Total RNA was prepared from wild-type N2 hermaphrodite populations synchronized at each of four (L1–L4) larval stages and at the adult (A) stage, from an N2 adult male population, and from fem-1(hc17lf), fem-3(q20gf), and glp-1(q231) adult hermaphrodites grown at 25°. Total RNA and cDNA were prepared and qPCR reactions were performed as previously described (Kim et al. 2010). The primers for act-1, which served as the internal control, were 5′-CCA GGA ATT GCT GAT CGT ATG CAG AA-3′ and 5′-TGG AGA GGG AAG CGA GGA TAG A-3′. The primers for cdc-25.4 were 5′-GAC AGG TAT CAG ACT AGA TTC TC-3′ and 5′-CAG CAC CCT TAA TAT GTC CA-3′. The relative expression level of each gene was defined as the mRNA level of each gene averaged from triplicate experiments and then normalized to that of act-1.
Male fertility test
To test cdc-25.4(tm4088) male fertility, we used fog-2(q71) mutants. Fog-2(q71) hermaphrodites are sterile owing to an absence of sperm (Schedl and Kimble 1988). For the fertility test, fresh NGM plates seeded with a 150 µl spot of OP50 overnight culture were prepared. A single L4 larval fog-2(q71) hermaphrodite and a single mutant male were placed on an OP50-spotted NGM plate and allowed to mate at 20°. They were transferred to a new OP50-spotted NGM plate every day for 3 d and the total number of outcrossed progeny was counted. As the negative and positive controls, single fog-2(q71) hermaphrodite-alone plates and single fog-2(q71) hermaphrodite mated with single him-5(e1467) male plates, respectively, were included in each experiment.
Immunofluorescence analysis
To observe the meiotic division processes during spermatogenesis, immunofluorescence analysis was performed as previously described (Kim et al. 2010). Briefly, adult virgin males were dissected and extruded gonads were fixed with cold methanol and 3% paraformaldehyde. The fixed specimens were incubated with 1:200-diluted FITC-conjugated anti-α-tubulin (mouse monoclonal, Sigma) and 1:200-diluted anti-phospho-Histone H3 (rabbit polyclonal, Upstate) as primary antibodies at 4° overnight. Alexa 546-conjugated donkey anti-rabbit IgG (1:500 dilution) (Molecular Probes) was used as the secondary antibody. The specimens were counterstained with 0.5 μg/ml Hoechst 33342 to visualize DNA.
In vitro sperm activation
In vitro sperm activation was carried out as previously described (Singaravelu et al. 2011) with some minor modifications. L4 males were isolated on OP50-seeded NGM plates and cultured on the plates at 20° for 2 d in the absence of hermaphrodites. Then, 10 of the virgin males were transferred to 7 μl of 1 × sperm medium (50 mM HEPES, 25 mM KCl, 45 mM NaCl, 1 mM MgSO4, 5 mM CaCl2, and 10 mM Dextrose; pH 7.8) with or without 2 mg/ml of pronase on a glass slide. Spermatids were released by cutting the tails. After incubating at RT for 5 min, a coverslip was gently overlaid and sealed with vaseline. Activation of spermatids to spermatozoa was observed at 630 × magnification under Nomarski differential interference contrast (DIC) microscopy.
Sperm transfer assay
To examine the transfer of sperm from a male to a hermaphrodite during mating, a single L4 male was mated with a single L4 fog-2(q71) hermaphrodite on a fresh OP50-spotted NGM plate at 20° for 2 d. After mating, the fog-2(q71) hermaphrodites were dissected, and extruded gonads were fixed with cold methanol and cold acetone. Then, the fixed gonads were immunostained with a sperm-specific mouse monoclonal antibody SP56 (a gift from Susan Strome), as previously described (Kim et al. 2010), to detect successfully transferred sperm in the gonads. Briefly, the fixed gonads were first incubated with 1:100-diluted SP56 at 4° overnight, followed by incubation with 1:500-diluted Alexa 488-conjugated goat anti-mouse IgG (Molecular Probes) secondary antibody at room temperature (RT) for 3 hr. DNA was counterstained with 0.5 µg/ml Hoechst 33342. For DNA staining of male gonads, virgin adult males were dissected, and extruded gonads were fixed with cold methanol and 3% paraformaldehyde. Then, the DNA in the fixed gonads was stained with 0.5 µg/ml Hoechst 33342.
Mating behavior assay
Observation of mating behavior was carried out as previously described (Liu et al. 2007). Before the assay, L4 males were isolated on an OP50-seeded NGM plate and incubated at 20° overnight. NGM plates were prepared 1 d prior to the experiments, and an OP50 overnight culture 3 mm in diameter was spotted on the plates. When the culture had been completely absorbed into the plates (after several hours), the OP50-spotted NGM plates were used for the following mating behavior assay: a single virgin 1-d-old adult male was placed on a mating plate that contained twenty evenly-distributed immotile unc-22(e66) adult hermaphrodites. Mating behavior was observed for 15 min or until ejaculation. Animals maintained at 20° were either observed at 65 × magnification under a Nikon SMZ1500 microscope or recorded at 63 × magnification using a Hamamatsu Orca ERG digital camera under a Zeiss Axioskop 2 MOT microscope with Nikon NIS-Elements Basic Research imaging software (version 4.3). Mating behavior was classified as previously described (Loer and Kenyon 1993; Koo et al. 2011). Percent contact response = (the number of successful contact responses/total number of male tail contacts with a hermaphrodite) × 100. Both tail contact and the initiation of backing are required for a successful contact response. Percent backing = (the number of completed backward locomotion/total number of backing attempts) × 100. Completed backward locomotion was defined as a male moving backward until reaching either the head or tail of a hermaphrodite. Turning behavior was classified and scored as previously described (Loer and Kenyon 1993). Completed turns consist of both good and sloppy turns. Percent vulva location and spicule insertion were also calculated by dividing the number of successful behaviors with total number of trials. Response time was measured from the time when a male was placed on a mating plate to the time of his first successful contact response.
Male tail-chasing behavior
Ten young adult males were transferred to a fresh NGM plate seeded with 10 µl of OP50. Male tail-chasing behavior was observed for 30 min at 20 × magnification. When a male circled at least once and touched his head to his tail, tail-chasing behavior was counted as one success.
Construction of transgenic reporter DNAs
All plasmid DNAs were constructed using the In-Fusion HD Cloning system (Clontech). A 6.8 kb genomic sequence spanning from 4.9 kb upstream from the cdc-25.4 start codon to 0.9 kb downstream from the cdc-25.4 stop codon was PCR-amplified from N2 genomic DNA using primer pairs specifically designed for the In-Fusion reaction. The amplified genomic sequence was ligated by an In-Fusion reaction with pCFJ352 vector DNA, which was linearized by double restriction enzyme digestion with SpeI and NcoI. Finally, a gfp coding sequence with artificial introns, which was PCR-amplified from the pJA256 vector, was inserted into the N-terminal end of the cdc-25.4 coding sequence to generate the Pcdc-25.4::gfp::cdc-25.4::cdc-25.4 3′ UTR translational reporter construct. The Pcdc-25.4::gfp transcriptional reporter construct was generated by removing the cdc-25.4 coding sequence from the translational reporter construct through inverse PCR and In-Fusion. Pmyo-3::mCherry::unc-54 3′ UTR (pCFJ104) was used to generate neuronal promoter-driven cdc-25.4 rescue constructs. First, pCFJ104 DNA was linearized by inverse PCR using primers designed at the end of Pmyo-3 and the beginning of unc-43 3′ UTR sequences. Then, a gfp::cdc-25.4 fusion sequence, which was PCR-amplified from the Pcdc-25.4::gfp::cdc-25.4::cdc-25.4 3′ UTR translational reporter construct, was inserted into the linearized pCFJ104 DNA by the In-Fusion reaction. The myo-3 promoter sequence was replaced with the promoter sequences of unc-119 (2.3 kb) (Maduro and Pilgrim 1995), tph-1 (3.2 kb) (Sze et al. 2000), tba-9 (2 kb) (Hurd et al. 2010), or pkd-2 (2 kb) (Barr and Sternberg 1999), which were PCR-amplified from N2 genomic DNA using specific primers. The vectors, pCFJ352, pJA256, pCFJ104, pGH8, and pCFJ90, were obtained from Addgene.
Generation of transgenic animals
Transgenic animals were generated using standard microinjection techniques (Mello and Fire 1995), except that the Pcdc-25.4::gfp::cdc-25.4::cdc-25.4 3′ UTR (10 ng/µl) construct was integrated into the ttTi4348 locus on chromosome I using the MosSCI technique (Frøkjær-Jensen et al. 2008). The DNA constructs were injected at the following concentrations: Pcdc-25.4::gfp::cdc-25.4 3′ UTR (20 ng/µl) with a coinjection marker, Prab-3::mCherry (10 ng/µl, pGH8); Punc-119::gfp::cdc-25.4::unc-54 3′ UTR (25 ng/µl), Ptph-1::gfp::cdc-25.4::unc-54 3′ UTR (50 ng/µl), Ptba-9::gfp::cdc-25.4::unc-54 3′ UTR (50 ng/µl), and Ppkd-2:gfp::cdc-25.4::unc-54 3′ UTR (50 ng/µl) with a coinjection marker; and Pmyo-2::mCherry (2 ng/µl, pCFJ90).
Microscopy
Transgenic worms were observed and recorded under an Olympus FV-1000 spectral confocal microscope with FV10-ASW 2.0 software. Specimens were observed and recorded under a Zeiss Axioskop 2 MOT fluorescence microscope with Openlab software (Improvision). Images were processed with Photoshop CC and Illustrator CC2015 software (Adobe).
Neurotransmitter treatment
Treatment with neurotransmitters was performed as previously described (Loer and Kenyon 1993; Sawin et al. 2000; LeBoeuf et al. 2014) with minor modifications. 100 mM serotonin creatinine sulfate monohydrate (5-HT, Sigma) and 200 mM dopamine hydrochloride (DA, Sigma) were freshly prepared in M9 buffer. 5-HT was dissolved in solution by heating at 65°. After autoclaved NGM agar medium had cooled, 5-HT and DA were added at 1 and 2 mM final concentrations, respectively. The NGM agar plates were dried at RT for several hours and OP50 overnight cultures with 1 mM 5-HT or 2 mM DA were spotted. The plates were allowed to dry at RT overnight in the dark. Virgin young adults were transferred to the plates containing neurotransmitters and incubated at 20° for 1–2 hr (5-HT) or 6–7 hr (DA). Turning behavior was observed as described previously.
RNA interference
RNAi was performed by the soaking method as previously described (Yoon et al. 2012). For PCR amplification of cDNA templates, the yk cDNA clone, yk321c10, for wee-1.3 was generously provided by Y. Kohara (National Institute of Genetics, Japan). cdc-25.4 cDNA was amplified from an N2 cDNA using gene-specific primers. Double-stranded (ds) RNA was transcribed in vitro from the PCR-amplified cDNA templates and purified as previously described (Maeda et al. 2001). Synchronized L1 worms were soaked in the dsRNA solution at 20° for 24 hr, allowed to recover on OP50-seeded NGM plates, and grown at 20° until they reached the adult stage.
Statistical analysis
All the experiments were repeated more than three times for statistical evaluation of data. P values were calculated by Student’s t-test. P < 0.05 was considered as statistically significant.
Data availability
Strains and reagents are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
cdc-25.4 is required for male fertility
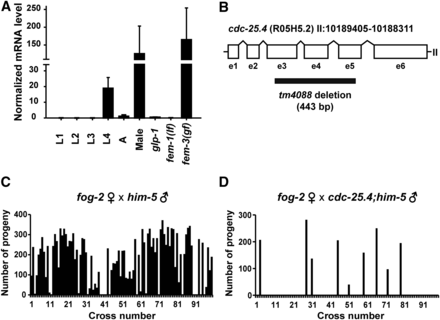
Expression levels of cdc-25.4 mRNA were examined at different larval stages in wild-type N2 hermaphrodites and in N2 adult males by qRT-PCR. cdc-25.4 mRNA was highly expressed in N2 adult males, and at the L4 larval stage, when maturation of reproductive organs actively occurs, in N2 hermaphrodites (Figure 1A). We also found that cdc-25.4 mRNA was abundantly expressed in fem-3(q20gf) hermaphrodites which produce only sperm (Barton et al. 1987), but not in glp-1(q231) hermaphrodites which contain few germ cells (Austin and Kimble 1987), or in fem-1(hc17lf) hermaphrodites which produce only oocytes (Nelson et al. 1978). These results suggest that cdc-25.4 should play a critical role in male reproduction.
cdc-25.4 is required for male reproduction. (A) cdc-25.4 mRNA (messenger RNA) expression levels were examined in larval (L1–L4) and adult (A) stages as well as in males, germline-proliferation-defective glp-1(q231) mutants, fem-1(hc17lf) mutants that produce only oocytes, and fem-3(q20gf) mutants that produce only sperm. The values were normalized to those of act-1. Error bars indicate SD. (B) Genomic structure of cdc-25.4 (R05H5.2), which is located on chromosome II. cdc-25.4 consists of six exons (e1–6) and five introns. The region of the tm4088 deletion (443 bp) is also shown. (C) The number of progeny produced by fog-2(q71) single hermaphrodites after mating with single him-5(e1467) males (n = 98). (D) The number of progeny produced by fog-2(q71) single hermaphrodites after mating with single cdc-25.4(tm4088); him-5(e1467) males (n = 97). Of cdc-25.4; him-5 males, 90.7% (88/97) failed to produce outcrossed progeny.
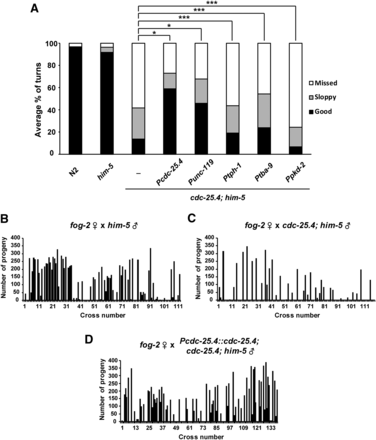
To study the roles of cdc-25.4, we first observed the phenotype of cdc-25.4(tm4088) deletion mutants, in which 443 bp, encompassing from exon 3 (e3) to exon 5 (e5), is deleted (Figure 1B). cdc-25.4(tm4088) mutant hermaphrodites are viable and fertile. Their brood size does not differ from that of wild-type hermaphrodites at 20° (Supplemental Material, Figure S1). To analyze the phenotype of cdc-25.4 mutant males, we introduced the him-5(e1467) mutation, which generates a substantial number of males among self-progeny (Hodgkin 1983), into the cdc-25.4 mutant background. First, we examined cdc-25.4(tm4088) male fertility to verify that cdc-25.4 functions in male reproduction. To test male fertility, a single fog-2(q71) hermaphrodite was mated with either a single him-5(e1467) male or a single cdc-25.4(tm4088); him-5(e1467) male, and the numbers of outcrossed progeny were scored. In fog-2(q71) hermaphrodites, the germline is completely feminized and spermatogenesis does not occur during the L4 larval stage (Schedl and Kimble 1988). Therefore, sperm must be provided by males for fog-2(q71) hermaphrodites to produce progeny. As expected, fog-2(q71) hermaphrodites incubated alone were all sterile (n = 30, data not shown). When fog-2(q71) hermaphrodites were mated with him-5(e1467) males, 87.8% of the fog-2(q71) hermaphrodite parents successfully produced outcrossed progeny (n = 98) (Figure 1C). However, when fog-2(q71) hermaphrodites were mated with cdc-25.4(tm4088); him-5(e1467) males, 90.7% of the fog-2(q71) hermaphrodite parents failed to produce outcrossed progeny (n = 97) (Figure 1D). Notably, several fog-2(q71) hermaphrodite parents that successfully produced outcrossed progeny after being mated with cdc-25.4; him-5 males generated similar numbers of outcrossed progeny as fog-2(q71) hermaphrodite parents that were mated with him-5 control males. This result suggests that, although the rate of successful mating was much lower than the control him-5 males, if mating was successful, cdc-25.4; him-5 males could transfer similar numbers of functional sperm to fog-2 hermaphrodites. Taken together, based on the results of expression analysis and male mating tests, we conclude that cdc-25.4 is required for either male mating behavior or male fecundity.
Sperm development is not defective in cdc-25.4 mutants
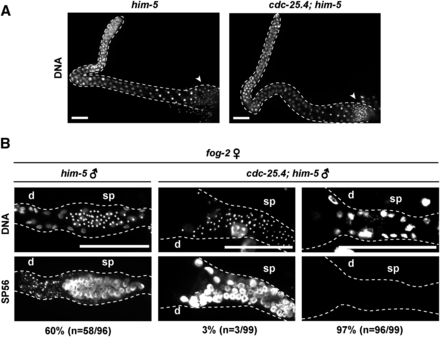
To determine whether sperm are produced normally in cdc-25.4 mutants, we examined male gonad arms after DNA staining. We found that the overall gonadal morphology of cdc-25.4(tm4088); him-5(e1467) males was indistinguishable from that of him-5(e1467) control males (Figure 2A), and that similar numbers of spermatids were observed in the proximal region of cdc-25.4; him-5 male gonads as in him-5 control male gonads (Figure 2A, arrowheads). Wild-type male gonad arms consist of two regions, the distal mitotic region and the proximal meiotic region. After germ cells proliferate in the distal region by mitotic division, they move to the proximal region and begin meiosis, and spermatogenesis occurs during the course of meiotic divisions I and II to produce spermatids. Finally, immotile spermatids are transformed into motile spermatozoa through a process called spermiogenesis or sperm activation (L’Hernault 2006). To determine whether germ cells in cdc-25.4; him-5 male gonads undergo normal spermatogenesis, we examined meiotic division processes during spermatogenesis by immunofluorescence analysis using an anti-α-tubulin antibody, which detects microtubules, and an anti-phospho histone H3 antibody, a metaphase marker. We found that meiotic divisions I and II occurred normally during spermatogenesis in cdc-25.4; him-5 male gonads as in him-5 control male gonads (Figure S2A). In C. elegans, male spermatids are activated when they are transferred to hermaphrodite uteri (Ward et al. 1983). Spermatids recovered from male gonads can be activated in vitro to become motile spermatozoa by treatment with several agents, including pronase (Ward et al. 1983). We found that spermatids obtained from cdc-25.4; him-5 male gonads were successfully activated in vitro after pronase treatment, as were spermatids from him-5 control male gonads (Figure S2, B and C). These results indicate that spermatogenesis (meiotic divisions) and spermiogenesis (sperm activation) occur normally in cdc-25.4 mutant male gonads, and thus, cdc-25.4 mutant males can produce functional sperm.
The majority of cdc-25.4(tm4088) males failed to transfer sperm to hermaphrodite uteruses. (A) DNA staining of male adult gonads. him-5(e1467) (n = 45) and cdc-25.4(tm4088); him-5(e1467) (n = 57) male adults were dissected and their gonads were stained with a DNA fluorescent dye, Hoechst 33342. Spermatids (arrowheads) were observed normally in the proximal region of cdc-25.4(tm4088); him-5(e1467) male gonads as compared with him-5(e1467) male gonads. Scale bars, 20 μm. (B) After mating with either single him-5(e1467) males or single cdc-25.4(tm4088); him-5(e1467) males, each fog-2(q71) hermaphrodite was dissected and the proximal region of the gonad was immunostained with a sperm-specific monoclonal antibody, SP56, along with Hoechst 33342 to visualize DNA. Scale bars, 50 μm. d, distal side of the gonad; sp, spermatheca.
cdc-25.4 is required for successful male mating
C. elegans males deliver their sperm to hermaphrodites through a complicated male-specific mating behavior. Since sperm production in cdc-25.4 mutant males was judged to be normal, we next asked whether the sperm of cdc-25.4 mutant males could be successfully transferred to hermaphrodites. To answer this question, we monitored sperm transfer by immunostaining fog-2(q71) hermaphrodite gonads with a sperm-specific monoclonal SP56 antibody after fog-2 hermaphrodites were mated with cdc-25.4 mutant males. We found that 97% of the fog-2 hermaphrodites failed to display sperm-specific SP56 signals after being mated with cdc-25.4; him-5 males, while 60% of the fog-2 hermaphrodites showed strong SP56 signals in their spermatheca after being mated with him-5 control males (Figure 2B). These results indicate that cdc-25.4 is required for successful sperm transfer, but not for sperm development or sperm activation. Therefore, when sperm were successfully transferred from a cdc-25.4 mutant male to a hermaphrodite, which occurred at a low frequency, that hermaphrodite could produce substantial numbers of outcrossed progeny (Figure 1D).
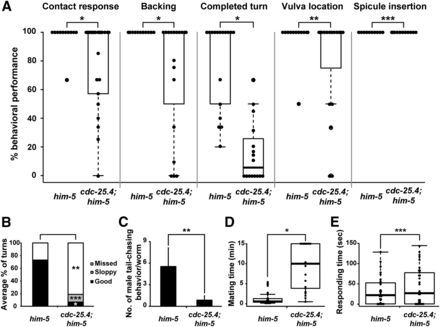
To further elucidate the roles of cdc-25.4 in male mating behavior, we attempted to determine in which step of mating behavior cdc-25.4 mutant males display a defect. Mating behavior can be subdivided into a series of processes, beginning with contact response, proceeding to backing, turning, and vulva location, then ending with spicule insertion (Loer and Kenyon 1993; Koo et al. 2011). We found that cdc-25.4; him-5 males showed defects in most of these steps except spicule insertion, compared with control him-5 males (Figure 3A). Among the mating processes, turning behavior was most severely affected in cdc-25.4 mutant males (P < 0.005). When defects in turning behavior were further divided into several classes according to the degree of the defect, we noticed that only 4.7% of the observed cdc-25.4; him-5 males showed “good” turning behavior (Figure 3B). That is, only 4.7% of tested cdc-25.4 mutant males could successfully turn without losing contact between their tails and the bodies of the hermaphrodites (movie File S1 and movie File S2). It was reported that appropriate tail curling is necessary for good male turning (Loer and Kenyon 1993). Tail curling is also required when a male makes a circular backward movement, which occurs when he chases his own head with his tail. This behavior is called “male tail-chasing behavior” (Wang et al. 2014). To determine whether cdc-25.4 mutant males show a defect in tail-chasing behavior, we scored how many times a cdc-25.4 mutant male successfully performed the tail-chasing behavior in a 30 min time interval (see Materials and Methods). We found that cdc-25.4; him-5 males performed significantly lower numbers of tail-chasing behavior compared to him-5 control males (P < 0.05) (Figure 3C). This result suggests that cdc-25.4 mutant males show a significant defect in turning behavior because their tails cannot curl well. We next measured the time required for mating. We defined this as a time interval required since each male was placed on a mating plate until he finished ejaculation. When mating did not occur or did not complete, we stopped the observation after 15 min elapsed, and considered the elapsed time of the trial to be 15 min. By this assay, we found that the time required for mating was significantly longer for cdc-25.4; him-5 males than for him-5 control males (P < 0.001) (Figure 3D). We also found that only when cdc-25.4; him-5 males contacted the ventral side of hermaphrodites by chance, could they locate the vulva and finish mating in a short period of time, possibly because they did not have to turn. By contrast, we found that responding time, which is the time interval since a male was placed on a mating plate until the male made his first successful contact with a hermaphrodite, was not significantly different between cdc-25.4; him-5 males and him-5 control males (P > 0.05) (Figure 3E). This result suggests that the longer mating time observed for cdc-25.4 mutant males was due to a defect that occurred after contacting hermaphrodites, most likely a defect in turning behavior. Taken together, these results indicate that the low fertility of cdc-25.4 mutant males was due to behavioral defects in several steps of the male mating processes.
cdc-25.4 functions in male mating behavior. (A) Percent behavioral performance in respective mating processes by him-5(e1467) males (n = 23) and by cdc-25.4(tm4088); him-5(e1467) males (n = 21). Male mating behavior was subdivided into the following processes: contact response, backing, completed turn, vulva location, and spicule insertion. Distribution of percent performance in the different processes displayed by each genotype is shown as a box plot. (B) Percent distribution of performance in turning behavior by him-5(e1467) males and by cdc-25.4(tm4088); him-5(e1467) males. Performance of turning behavior was classified into three levels: good, sloppy, and missed. (C) cdc-25.4(tm4088) mutant males showed defective tail-chasing behavior. The number of successful male tail-chasing incidents was scored in him-5(e1467) males (n = 17) and cdc-25.4(tm4088); him-5(e1467) males (n = 30) for 30 min. (D) Distribution of the time period (min) required for each mating event by him-5(e1467) males and by cdc-25.4(tm4088); him-5(e1467) males, displayed as a box plot. (E) Distribution of the time (sec) required to respond to hermaphrodites by him-5(e1467) males and by cdc-25.4(tm4088); him-5(e1467) males is displayed as a box plot. Box plots were created using R. Horizontal lines represent minimum, lower quartile (25%), median (50%), upper quartile (75%), and maximum, from the bottom to the top, in the box plot. Outliers represent less or more than two-thirds of the quartile. Dot indicates individual data points. Error bars indicate SD. P values were calculated by Student’s t-test against him-5 controls. * P < 0.005, ** P < 0.05, and *** P > 0.05.
cdc-25.4 is expressed in many neuronal cells and germ cells
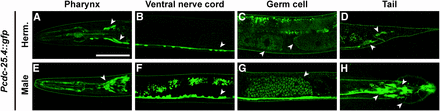
To determine the expression pattern of cdc-25.4, we generated several lines of transgenic animals carrying a Pcdc-25.4::gfp reporter construct. Surprisingly, we found that Pcdc-25.4::gfp was expressed in many neurons, including pharyngeal neurons (Figure 4, A and E), the ventral nerve cord (Figure 4, B and F), and tail neurons (Figure 4, D and H) in both hermaphrodites and males. However, the signal was not detected in the dorsal nerve cord. Pcdc-25.4::gfp expression was also observed in pharyngeal muscles, the head ganglion, preanal ganglion, and lumbar ganglion. In particular, Pcdc-25.4::gfp was expressed in ray cell bodies, spicule neurons, hook neurons, and postcloacal sensilla neurons in male tails (Figure S3). cdc-25.4 was also expressed in germ cells, including oocytes and sperm (Figure 4, C and G). We also found that Pcdc-25.4::gfp was expressed in many neurons throughout larval development (Figure S4). Neuronal expression of cdc-25.4 was confirmed by colocalization of a pan-neuronal marker, Prab-3::mCherry (Figure S4).
Pcdc-25.4::gfp is expressed in many neurons and germ cells. The Pcdc-25.4::gfp transgene was expressed in the pharynx (A, E), ventral nerve cord (B, F), oocytes (C), sperm (C, G), and tail neurons (D, H), in the transgenic hermaphrodites (Herm.) and males. Arrowheads indicate the loci where Pcdc-25.4::gfp was detected. Scale bar, 50 μm.
Turning defect of cdc-25.4 mutant males was rescued by pan-neuronal expression of a cdc-25.4 transgene
To test whether neuronal expression of a cdc-25.4 transgene can rescue the defect in turning behavior seen in cdc-25.4 mutant males, we generated several cdc-25.4 transgene constructs in which different promoters controlled the expression of cdc-25.4. These cdc-25.4 transgenes were examined to see which promoter-driven transgene could successfully rescue the turning defect of cdc-25.4 mutant males. First, we confirmed that cdc-25.4’s own promoter-driven cdc-25.4 transgene significantly rescued the turning defect of cdc-25.4 mutant males (P < 0.001) (Figure 5A). The fertility defect of cdc-25.4 mutant males was also rescued by cdc-25.4 promoter-driven cdc-25.4 transgene expression (Figure 5, B–D). Furthermore, we found that a cdc-25.4 transgene driven by the unc-119 promoter, which is expressed pan-neuronally, could significantly rescue the turning defect of cdc-25.4; him-5 males (P < 0.001) (Figure 5A), indicating that cdc-25.4 is required in neurons for successful male turning.
The rescue of defective phenotypes in cdc-25.4(tm4088) males. (A) The turning behavior in cdc-25.4(tm4088) males was rescued by a pan-neuronal promoter-driven cdc-25.4 transgene. Possible rescue of defective turning behavior in cdc-25.4(tm4088) males by several neuron-specific promoter-driven cdc-25.4 transgenes was tested. Levels of turning behavior were classified into good, sloppy, and missed. Percent distribution of the respective turning behavior levels were measured in N2 (n = 21), him-5(e1467) (n = 59), and cdc-25.4(tm4088); him-5(e1467) (n = 61) males, as well as in cdc-25.4(tm4088); him-5(e1467) males harboring a gfp::cdc-25.4 transgene driven by either the cdc-25.4 (positive control, n = 21), unc-119 (expressed pan-neuronally, n = 14), tph-1 (expressed in serotonergic neurons, n = 21), tba-9 (expressed in ray A neurons, n = 20), or pkd-2 (expressed in ray B neurons, n = 8) promoters. (B) The number of progeny produced by fog-2(q71) single hermaphrodites after mating with single him-5(e1467) males (n = 113). 64.6% of the fog-2(q71) hermaphrodite parents produced outcrossed progeny. (C) The number of progeny produced by fog-2(q71) single hermaphrodites after mating with single cdc-25.4(tm4088); him-5(e1467) males (n = 119). 68.1% of the fog-2(q71) hermaphrodite parents failed to produce outcrossed progeny. (D) The number of progeny produced by fog-2(q71) single hermaphrodites after mating with single Pcdc-25.4::cdc-25.4; cdc-25.4(tm4088); him-5(e1467) males (n = 140). 54.3% of the fog-2(q71) hermaphrodite parents produced outcrossed progeny. P values were calculated by Student’s t-test. * P < 0.001 and *** P > 0.05.
It is known that male turning behavior, the most prominent defective phenotype in cdc-25.4 mutant males, is mediated by 5-HT and DA (Loer and Kenyon 1993; Liu and Sternberg 1995). Exogenous 5-HT treatment led to an increase in male tail curling behavior (Loer and Kenyon 1993). In addition, ablation of serotonergic CP motor neurons, which are located in the male ventral nerve cord, and 5-HT-deficient mutant males exhibited reduced turning behavior (Loer and Kenyon 1993). In addition, the R5A, R7A, and R9A neurons, which release DA, are required for tail ventral turning, and ablation of these neurons in males results in sloppy turning behavior (Sulston et al. 1975; Sulston and Horvitz 1977; Liu and Sternberg 1995). Based on these previous studies, we questioned whether cdc-25.4 is involved in either synthesis or secretion of these neurotransmitters. To test this possibility, cdc-25.4 mutant males were exogenously treated with neurotransmitters. However, we found that exogenous DA failed to rescue the turning defect of cdc-25.4; him-5 males (Figure S5A). Furthermore, 5-HT treatment also failed to rescue the turning defect of cdc-25.4; him-5 males (Figure S5B). These results suggest either that cdc-25.4 does not have an essential function in serotonergic or dopaminergic neurons or that cdc-25.4 is not required for the synthesis or secretion of these neurotransmitters.
Although exogenous treatment of 5-HT and DA failed to rescue the turning defect of cdc-25.4 mutant males, to confirm that cdc-25.4 does not have an essential function in serotonergic or dopaminergic neurons, we tested whether a cdc-25.4 transgene driven by the tph-1 promoter or the dat-1 promoter could rescue the turning defect of cdc-25.4 mutant males. tph-1 and dat-1 are specifically expressed in serotonergic and dopaminergic neurons, respectively. tph-1 encodes a tryptophan hydroxylase that is required for 5-HT biosynthesis (Sze et al. 2000). dat-1 encodes a DA transporter and controls dopaminergic neurotransmission (Jayanthi et al. 1998; Nass et al. 2002). We found that expression of a cdc-25.4 transgene under the control of the tph-1 promoter could not rescue the turning defect of cdc-25.4 mutant males to a significant degree (P > 0.05). This result suggests that expression of cdc-25.4 in serotonergic neurons alone is not sufficient to rescue the cdc-25.4 male mutant phenotype (Figure 5A). Because we failed to obtain transgenic animals in which a cdc-25.4 transgene is expressed under the control of the dat-1 promoter, we could not determine whether expression of cdc-25.4 in dopaminergic neurons alone is sufficient to rescue the cdc-25.4 male mutant phenotype. We suspect that cdc-25.4 expression under the control of the dat-1 promoter may induce some toxic effects. In place of the dat-1 promoter, we tried to express cdc-25.4 transgenes under the control of the tba-9 promoter and the pkd-2 promoter to express cdc-25.4 in ray A-type and B-type neurons, respectively. tba-9 is one of the nine α tubulins and is expressed in various ciliated sensory neurons, including several male-specific neurons (Gogonea et al. 1999; Hurd et al. 2010). pkd-2 is expressed in male-specific sensory neurons and required for male mating behavior (Barr and Sternberg 1999; Barr et al. 2001). We found that cdc-25.4 transgene expression in ray A-type neurons under the control of the tba-9 promoter rescued the turning defect of cdc-25.4 mutant males slightly, but not significantly (P > 0.05) (Figure 5A). It must be noted that tba-9 expression is not limited to ray A-type neurons. It is also expressed in other ciliated neurons that are located in the head and ventral nerve cord (Hurd et al. 2010). The tba-9 promoter-driven cdc-25.4 expression in multiple ciliated neurons might have contributed to the slight rescue of the defective turning behavior. By contrast, cdc-25.4 transgene expression under the control of the pkd-2 promoter completely failed to rescue the turning defect of cdc-25.4 mutant males. These results suggest that cdc-25.4 would rather function in multiple neurons than a subset of serotonergic, dopaminergic, or ray neurons.
The neuronal morphology of cdc-25.4 mutant males is generally normal
Since cdc-25.4 is expressed in many neurons and considered to be an evolutionarily conserved cell division cycle regulator, we questioned whether cdc-25.4 affects neuronal cell divisions in males. If neuronal cell divisions in cdc-25.4 mutant males are defective, this defect most likely results in abnormal numbers of neurons, and thus, abnormal neuronal morphology. To test this possibility, we compared gross neuronal morphology between him-5 control adult males and cdc-25.4; him-5 adult males under fluorescence microscopy after introducing a pan-neuronally expressing Punc-119::gfp transgene to these males (Figure S6). Despite careful examination, we could not detect any obvious difference in the neuronal expression pattern of the Punc-119::gfp transgene between him-5 control males and cdc-25.4; him-5 males.
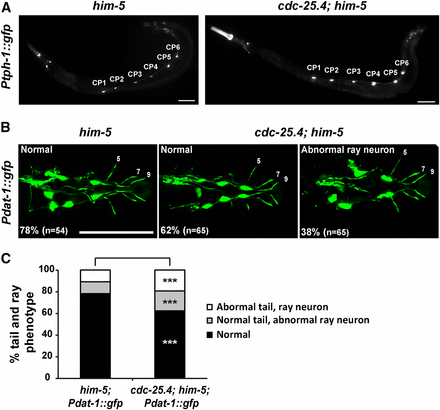
Although cdc-25.4 transgene expression in serotonergic and dopaminergic neurons under the control of the tph-1 promoter and the dat-1 promoter, respectively, could not rescue the turning defect of cdc-25.4 mutant males, to determine whether cdc-25.4 mutant males have any morphological defects in these neurons, we examined the morphology of serotonergic CP motor neurons and dopaminergic R5A, R7A, and R9A neurons after introducing Ptph-1::gfp and Pdat-1::gfp transgenes into cdc-25.4 mutant males. We found that CP 1-6 motor neurons were morphologically normal in cdc-25.4; him-5 males as compared to him-5 control males (Figure 6A). On the other hand, we found that cdc-25.4; him-5 males showed slight abnormalities in ray neuronal morphology (38%, n = 65) as compared to him-5 control males (22%, n = 54), although the difference was not significant (P > 0.05) (Figure 6, B and C). These results suggest that the defect in turning behavior in cdc-25.4 mutant males is most likely not caused by a loss of neurons or abnormal neuronal morphology.
The number and morphology of serotonergic CP neurons and dopaminergic ray neurons appear normal in cdc-25.4 mutant males. (A) Male-specific CP1–6 neurons, which were labeled with the Ptph-1::gfp transgene, were detected normally on the ventral nerve cord in both him-5(e1467) and cdc-25.4(tm4088); him-5(e1467) males. (B) The number and morphology of dopaminergic ray neurons (R5A, R7A, and R9A), which were labeled with the Pdat-1::gfp transgene, were not significantly different between him-5(e1467) and cdc-25.4(tm4088); him-5(e1467) males. (C) Percent distribution of male tail phenotypes in him-5(e1467); Pdat-1::gfp and cdc-25.4(tm4088); him-5(e1467); Pdat-1::gfp transgenic males. Male tail phenotypes were classified into three classes: normal tail morphology with normal DA ray neurons, normal tail morphology with abnormal DA ray neurons, and abnormal tail morphology with abnormal DA ray neurons. Scale bars, 50 μm. P values were calculated by Student’s t-test against him-5; Pdat-1::gfp controls. *** P > 0.05. DA, dopamine.
cdc-25.4 may function in a noncanonical manner in male neurons to regulate male mating
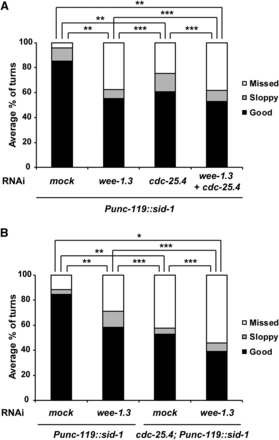
In canonical cell division cycle regulation, Wee1 kinase and Cdc25 phosphatase compete against each other to regulate Cdk1 activity (Fantes 1979). That is, Wee1 kinase inactivates Cdk1 by phosphorylating specific amino acid residues on Cdk1, and Cdc25 phosphatase activates Cdk1 by removing inhibitory phosphates from these residues (Hagan and Grallert 2013). We previously reported that cdc-25.1 and cdc-25.2, members of the C. elegans cdc-25 family, counteract wee-1.3 in germ cell proliferation, oocyte maturation, and intestinal division (Kim et al. 2010; Yoon et al. 2012; Lee et al. 2016). Three wee-1 genes have been identified in C. elegans; wee-1.1, wee-1.2, and wee-1.3. wee-1.1 is only expressed in the E cell of the 12-cell-stage embryo, which develops into the intestine. wee-1.2 is a pseudogene that is not transcribed (Wilson et al. 1999; Robertson et al. 2014). wee-1.3 is expressed in the embryos, adult soma, and germline (Lamitina and L’Hernault 2002; Allen et al. 2014). To determine whether wee-1.3 also counteracts cdc-25.4 in neurons during male mating, we examined the male turning behavior of cdc-25.4 mutants with or without RNAi depletion of wee-1.3. We found that wee-1.3 RNAi did not suppress the turning defect of cdc-25.4 mutant males (Figure S7A). Instead, we found that wee-1.3 RNAi single-handedly caused a severe turning defect in him-5 control males. Furthermore, mating time was significantly extended after wee-1.3 RNAi treatment in him-5 control males (Figure S7B). We also observed that rays were abolished in male tails when wee-1.3 alone was depleted by RNAi (Figure S7C). Depletion of wee-1.3 caused severe defects not only in neuronal cells, but also in other types of cells. These cellular defects might be the cause of defective male mating in wee-1.3 RNAi-treated animals. To determine the function of wee-1.3 in neurons, we performed neuron-specific RNAi depletion of wee-1.3 using a strain, TU3311: Punc-119::sid-1, in which neuronal RNAi is specifically enhanced. This strain expresses the sid-1(+) transgene in all the neuronal cells by the unc-119 promoter (Calixto et al. 2010), which results in enhancement of neuronal RNAi. In addition, pan-neuronal expression of sid-1 reduces RNAi efficiency in nonneuronal tissues. We found that RNAi depletion of wee-1.3 in Punc-119::sid-1 worms caused significant defects in male turning behavior (Figure 7A). On the other hand, this neuron-specific wee-1.3 RNAi did not cause any significant defects in male tail development (Figure S7C), suggesting that wee-1.3 specifically functions in neuronal cells to control male mating behavior. We also confirmed that neuron-specific cdc-25.4 RNAi in Punc-119::sid-1 worms caused significant defects only in male turning behavior (Figure 7A). However, double RNAi depletion of wee-1.3 and cdc-25.4 in Punc-119::sid-1 worms did not suppress the turning defects observed in the cdc-25.4 single RNAi-treated Punc-119::sid-1 worms (Figure 7A). To confirm the relationship between cdc-25.4 and wee-1.3 in mating behavior, we introduced cdc-25.4 deletion mutation into the Punc-119::sid-1 worms, and tested the turning behavior after wee-1.3 RNAi treatment. We found that neuron-specific wee-1.3 RNAi did not suppress the turning defects of cdc-25.4 mutant males (Figure 7B), suggesting that wee-1.3 and cdc-25.4 are not competing against each other for the control of male turning behavior. Therefore, cdc-25.4 and wee-1.3 likely function independently for the control of male mating. These results also imply that, unlike other cdc-25 family members, cdc-25.4 may play a noncanonical role in neurons, which is distinct from canonical cell cycle regulation.
RNAi depletion of wee-1.3 did not suppress defective turning behavior of cdc-25.4(tm4088) males. (A) Levels of turning behavior in Punc-119::sid-1 neuron-specific RNAi males, which were classified into good, sloppy, and missed, were measured after either mock (n = 16), wee-1.3 (n = 18), cdc-25.4 (n = 22), or wee-1.3 plus cdc-25.4 double (n = 13) RNAi depletion at the L1 larval stage. (B) Levels of turning behavior in Punc-119::sid-1 and cdc-25.4(tm4088); Punc-119::sid-1 males were measured after either mock (n = 18) or wee-1.3 (n = 19) RNAi depletion at the L1 larval stage. P values were calculated by Student’s t-test against mock RNAi. * P < 0.001, ** P < 0.05, and *** P > 0.05. RNAi, RNA interference.
Discussion
In this study, we found that cdc-25.4 is required for male fertility through controlling the mating behavior. It appears that cdc-25.4 does not function only in a single step during the mating processes, because cdc-25.4 mutant males showed behavioral defects in various processes, including contact response, backing, turning, and vulva location, but not in spicule insertion (Figure 3A). In addition, cdc-25.4 is expressed in many neuronal cells, like the pan-neuronal gene rab-3 (Figure S4). Transgenic expression of cdc-25.4, driven either by its own promoter or by the pan-neuronal unc-119 promoter, rescued the cdc-25.4 mutant males’ mating ability. However, cdc-25.4 transgenes under the control of promoters that are expressed in specific types of neurons failed to rescue the cdc-25.4 mutant male phenotypes (Figure 5A). These results suggest that cdc-25.4 has multiple functions in neurons and that it is required pan-neuronally for male fertility. It took much longer time periods for the mutant males to locate vulva and ejaculate sperm, because cdc-25.4 mutant males are defective in contact response, backing, and turning, which are necessary for finding vulva and ejaculating sperm (Figure 3, A–D). However, response time to hermaphrodites was not delayed in cdc-25.4 mutant males, suggesting that they are not defective in searching for hermaphrodites (Figure 3E).
Although the cdc-25.4 mRNA expression level was high in the L4 larval stage in hermaphrodites, when spermatogenesis occurs, and Pcdc-25.4::gfp was expressed in sperm, neither spermatogenesis nor spermiogenesis were impaired in both cdc-25.4 mutant males and hermaphrodites (Figure S2). Unexpectedly, we did not observe any obvious defects in hermaphrodite or male germlines, suggesting that the other members of the cdc-25 gene family compensate for the function of cdc-25.4 in the germline, unlike in neuronal cells. All four members of the C. elegans cdc-25 gene family, cdc-25.1 (Ashcroft et al. 1999; Ashcroft and Golden 2002; Kim et al. 2009; Yoon et al. 2012), cdc-25.2 (Kim et al. 2010), cdc-25.3 (unpublished data), and cdc-25.4 (Figure 1A), are highly expressed in the germline. Therefore, they are expected to have functions in the germline. However, unlike other family members, we did not observe any obvious germline defects in cdc-25.4 mutants. Considering the expression patterns of the cdc-25 gene family, possible germline function of cdc-25.4 can be redundant with that of cdc-25.1 in the male germline (Ashcroft et al. 1999), and with that of cdc-25.1, cdc-25.2, and cdc-25.3 in the hermaphrodite germline. This possibility remains to be elucidated. While the expression level of cdc-25.4, which was quantified by qRT-PCR, suggested that cdc-25.4 is abundantly expressed in the germline (Figure 1A), the cdc-25.4 transgene driven by its own promoter was mainly expressed in neuronal cells including the male-specific neurons. It is known that male-specific neurons are formed during the L3 and L4 larval stages by growing axons and dendrites (Sulston et al. 1980). The fact that the highest expression of cdc-25.4 was observed at the L4 larval stage supports the idea that activity of cdc-25.4 is required for male neuronal cell functions.
The roles of C. elegans cdc-25 family genes in cell cycle progression in the germline (Ashcroft et al. 1999; Ashcroft and Golden 2002; Kim et al. 2009, 2010; Yoon et al. 2012), and in the intestine (Hebeisen and Roy 2008; Segref et al. 2010; Lee et al. 2016) have been previously reported. However, in this study, we revealed a noncanonical function of cdc-25.4 that is uncoupled from cell cycle regulation, based on the following three observations. (1) neuron-specific depletion of wee-1.3, a C. elegans ortholog of Wee1/Myt1 kinase, in cdc-25.4 mutant males did not suppress the defective phenotypes of cdc-25.4 mutant males. WEE-1.3 inactivates cyclin-dependent kinases by adding inhibitory phosphates, which are removed by CDC-25 phosphatase to promote the cell cycle (Fantes 1979; Russell and Nurse 1987). Thus, wee-1.3 normally counteracts cdc-25 in canonical cell cycle regulation. (2) The expression patterns of the Punc-119::gfp transgene in neuronal cells were indistinguishable between cdc-25.4 mutants and the wild type. (3) cdc-25.4 was expressed in already differentiated postmitotic neurons. These observations suggest that cdc-25.4 is required for neuronal function rather than neuronal development. Noncanonical functions of some cell cycle regulators have been previously reported. CDK7, CDK8, and CDK9 regulate transcription in humans (Malumbres 2014). Cell cycle-independent roles of CDK5 have been identified in a mammalian nervous system (Su and Tsai 2011). Interestingly, C. elegansCDK-5 is also required for neuronal functions (Ou et al. 2010; Park et al. 2011; Goodwin et al. 2012; Monteiro et al. 2012). Further, C. elegansCDK-8 regulates axon navigation decisions in the nervous system (Steimel et al. 2013). These studies support the idea that functions of cell cycle regulators are not limited to the cell cycle regulation. We should rather consider that they have the potential to execute noncanonical functions.
The most prominent behavioral defect observed in cdc-25.4 mutant males was the turning defect. Previous studies indicate that the male-specific CP ventral cord motor neurons (Loer and Kenyon 1993) and the three posterior-most rays, rays 7–9, are involved in male turning behavior (Liu and Sternberg 1995). Ablation of CP and ray 7–9 neurons caused defects in turning behavior (Loer and Kenyon 1993; Liu and Sternberg 1995). The CP neurons control tail moving by forming synapses with the male-specific diagonal muscles (White 1988). 5-HT and DA neurotransmitters were previously reported to be involved in male turning behavior (Loer and Kenyon 1993; Liu and Sternberg 1995). However, we found that the defective turning behavior in cdc-25.4 mutant males was not caused by a failure in the synthesis or secretion of these neurotransmitters, because cdc-25.4 mutant phenotypes were not recovered when we treated the worms with 5-HT or DA (Figure S5). Furthermore, CP neurons and R5A, R7A, and R9A neurons visualized by Ptph-1::gfp and Pdat-1::gfp expression were morphologically normal in cdc-25.4 mutant males. This observation suggests that cdc-25.4 is not involved in the formation of these neurons (Figure 6). In summary, we identified a novel noncanonical function of CDC-25.4 phosphatase in neuronal cells that is required for successful male mating behavior.
Acknowledgments
We thank Y. Kohara (National Institute of Genetics, Japan) for the yk cDNA clone, S. Strome (University of California, Santa Cruz, CA) for providing the SP56 antibody, S. Lee (Hanyang University, South Korea) for discussion, J.H.S. (Konkuk University, South Korea) for technical support, and H. Jung in J. Ahnn’s lab (Hanyang University, South Korea) for technical advice. We also thank S. Mitani and the National Bioresource Project (Japan) for providing cdc-25.4(tm4088). Some strains were provided by the Caenorhabditis elegans Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This research was supported by a Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology to Y.H.S. (2013R1A1A2009090 and 2015R1D1A1A01057488) and I.K. (2013 R1A1A2009820 and 2015R1D1A1A01057853).
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.036129/-/DC1.
Communicating editor: S. Lee
Literature Cited
Barr, M. M., and L. R. Garcia, 2006 Male mating behavior (June 19, 2006). WormBook, ed. The C. elegans Research Community WormBook, doi:10.1895/wormbook.1.78.1, http://www.wormbook.org.
L’Hernault, S. W., 2006 Spermatogenesis (February 20, 2006) WormBook, ed. The C. elegans Research Community WormBook, doi:10.1895/wormbook.1.85.1, http://www.wormbook.org.