-
PDF
- Split View
-
Views
-
Cite
Cite
Kyle Rhodehouse, Katherine Cascino, Laura Aseltine, Allegra Padula, Rachel Weinstein, Joseph S Spina, Christiane E Olivero, Priscilla M Van Wynsberghe, The Doubletime Homolog KIN-20 Mainly Regulates let-7 Independently of Its Effects on the Period Homolog LIN-42 in Caenorhabditis elegans, G3 Genes|Genomes|Genetics, Volume 8, Issue 8, 1 August 2018, Pages 2617–2629, https://doi.org/10.1534/g3.118.200392
Close - Share Icon Share
Abstract
The Caenorhabditis elegans (C. elegans) heterochronic pathway, which regulates developmental timing, is thought to be an ancestral form of the circadian clock in other organisms. An essential member of this clock is the Period protein whose homolog, lin-42, in C. elegans is an important heterochronic gene. LIN-42 functions as a transcriptional repressor of multiple genes including the conserved lin-4 and let-7 microRNAs. Like other Period proteins, levels of LIN-42 oscillate throughout development. In other organisms this cycling is controlled in part by phosphorylation. KIN-20 is the C. elegans homolog of the Drosophila Period protein kinase Doubletime. Worms containing a large deletion in kin-20 have a significantly smaller brood size and develop slower than wild type C. elegans. Here we analyze the effect of kin-20 on lin-42 phenotypes and microRNA expression. We find that kin-20 RNAi enhances loss-of-function lin-42 mutant phenotypes and that kin-20 mutant worms express lower levels of LIN-42. We also show that kin-20 is important for post-transcriptional regulation of mature let-7 and lin-4 microRNA expression. In addition, the increased level of let-7 found in lin-42(n1089) mutant worms is not maintained after kin-20 RNAi treatment. Instead, let-7 is further repressed when levels of kin-20 and lin-42 are both decreased. Altogether these results suggest that though kin-20 regulates lin-42 and let-7 microRNA, it mainly affects let-7 microRNA expression independently of lin-42. These findings further our understanding of the mechanisms by which these conserved circadian rhythmic genes interact to ultimately regulate rhythmic processes, developmental timing and microRNA biogenesis in C. elegans.
Many organisms exhibit circadian rhythmic behaviors that cycle in accordance with the Earth’s rotation and thus exhibit periodicities of ∼24 hr. These circadian rhythms can be entrained by environmental stimuli and persist in the absence of such cues, are unchanged by variations in temperature, and can be reset by external stimuli. Other fundamental aspects of life are governed by ultradian or infradian rhythms with periods that are either less or greater than the canonical 24 hr circadian rhythm, respectively. Central to these rhythms is the circadian clock, a group of highly conserved genes that function in oscillating feedback loops that are controlled by both transcriptional and post-transcriptional mechanisms.
In the nematode C. elegans, locomotion, resistance to osmotic stress, melatonin biosynthesis, and multiple metabolic variables can be entrained by light cues with a daily periodicity (Kippert et al. 2002; Saigusa et al. 2002; Migliori et al. 2011, 2012). Other rhythmic behaviors in C. elegans, like olfactory response, have been entrained by alterations in temperature (Olmedo et al. 2012). C. elegans also exhibits other essential ultradian rhythms like defecation (Iwasaki et al. 1995; Kobayashi et al. 2011). Though C. elegans do not express many of the proteins found in the classical circadian clocks of mammals and Drosophila, they do express many core clock proteins (Banerjee et al. 2005; Romanowski et al. 2014). Interestingly, many of these core clock proteins also function in the heterochronic pathway of C. elegans that regulates developmental timing (Banerjee et al. 2005; Temmerman et al. 2011). Thus the heterochronic pathway has been hypothesized to be an ancestral form of the circadian clock system (Temmerman et al. 2011).
Like the core circadian clock, the heterochronic pathway consists of many genes that successively regulate one another through feedback mechanisms to ultimately promote C. elegans development through four larval stages and into adulthood (Rougvie and Moss 2013). Proper progression through the heterochronic pathway can be assessed by evaluating the development of heterochronic seam cells (Resnick et al. 2010). Specific numbers of seam cells are present during each larval stage, and after the L4-to-adult molt, seam cells exit the cell cycle, fuse and generate a ridged cuticle structure called alae (Resnick et al. 2010). Thus, heterochronic mutants often display altered seam cell numbers and/or precocious or retarded alae formation (Resnick et al. 2010).
Based on its sequence similarity and rhythmic expression, the heterochronic gene lin-42 is the C. elegans homolog of the Period protein (Jeon et al. 1999). Period proteins act as transcriptional repressors to regulate circadian rhythms (Hardin 2005; Yu and Hardin 2006). LIN-42 also acts as a transcription factor to regulate expression of a multitude of genes (Mcculloch and Rougvie 2014; Van Wynsberghe and Pasquinelli 2014; Van Wynsberghe et al. 2014; Perales et al. 2014). In addition, lin-42 is essential for proper developmental timing, molting, and entry into an alternative dauer larval stage (Tennessen et al. 2006, 2010; Monsalve et al. 2011; Edelman et al. 2016). Consequently, lin-42 mutant worms show defects in circadian rhythmic activity, and exhibit a dumpy phenotype and precocious alae formation (Abrahante et al. 1998; Simonetta et al. 2009; Van Wynsberghe and Pasquinelli 2014).
Oscillation of Period is controlled both transcriptionally and post-transcriptionally. One protein important for this regulation is the kinase Doubletime in Drosophila and Casein Kinase 1ε and δ (CKIε/δ) in mammals (Kloss et al. 1998; Price et al. 1998). Phosphorylation by Doubletime decreases the stability and thus the levels and subcellular localization of Period (Price et al. 1998; Cyran 2005; Crane and Young 2014). The C. elegans homolog of Doubletime and CK1ε/δ is KIN-20 (Banerjee et al. 2005). RNAi against kin-20 causes some precocious developmental timing defects (Banerjee et al. 2005), though the reason for these defects and the effect of KIN-20 on the Period protein homolog LIN-42 is unknown.
Some of the genes associated with the heterochronic pathway function as small RNAs called microRNAs (miRNAs) (Rougvie and Moss 2013). miRNAs post-transcriptionally regulate gene expression by binding imperfectly to target mRNAs to ultimately inhibit their expression (Finnegan and Pasquinelli 2013). LIN-42 negatively regulates a broad range of miRNAs including let-7 and lin-4 (Mcculloch and Rougvie 2014; Van Wynsberghe et al. 2014; Perales et al. 2014). The conserved miRNA let-7 is essential for promoting cellular differentiation later in development (Resnick et al. 2010; Mondol and Pasquinelli 2012; Lee et al. 2016). Accordingly, under-expression of let-7 is associated with retarded development and a bursting vulva phenotype in C. elegans, as well as breast, lung and colon cancer in humans (Sayed and Abdellatif 2011; Mondol and Pasquinelli 2012; Lee et al. 2016). Loss of the conserved lin-4 microRNA, which is first expressed during the first larval stage, also causes retarded development in C. elegans (Ambros and Horvitz 1984; Ambros 1989).
miRNAs like let-7 and lin-4 ultimately function as ∼22 nucleotide (nt) RNAs, however they are initially transcribed from the genome by RNA polymerase II into long, primary miRNAs (pri-miRNAs) that are subsequently capped and polyadenylated (Lee et al. 2004). These primary miRNAs are then processed by the Microprocessor complex, which is composed of the RNase III enzyme Drosha and the RNA binding protein DGCR8 (also known as Pasha), into a ∼70 nt hairpin structured precursor miRNA (pre-miRNA) (Resnick et al. 2010; Finnegan and Pasquinelli 2013). Following export to the cytoplasm, the pre-miRNA is further processed by a second RNase III enzyme Dicer into the ∼22 nt mature miRNA (Resnick et al. 2010; Finnegan and Pasquinelli 2013). This mature miRNA is loaded onto Argonaute to form the miRNA-induced silencing complex (miRISC) (Resnick et al. 2010; Finnegan and Pasquinelli 2013). miRISC then uses the miRNA as a guide to bind and downregulate target gene expression (Resnick et al. 2010; Finnegan and Pasquinelli 2013). Consequently, aberrant levels of the mature miRNA can cause inappropriate expression of target genes and thus subsequent phenotypic effects. To ensure proper miRNA expression, each step in miRNA biogenesis is subject to regulation (Finnegan and Pasquinelli 2013). Some regulators act on a specific miRNA, while others act more globally to control miRNA expression (Finnegan and Pasquinelli 2013; Lee et al. 2016).
Here we utilized a large deletion of kin-20, kin-20(ok505), to analyze the effect of kin-20 on organismal development, lin-42 phenotypes and expression, and let-7 and lin-4 miRNA biogenesis. We found that kin-20 mutants have significantly fewer progeny and a slower growth than WT N2 worms, though they do not exhibit classical developmental timing defects in alae production. Because kin-20(ok505);lin-42(n1089) worms were lethal, we analyzed the impact of kin-20 RNAi on lin-42(n1089) mutant worms. We found that under-expression of kin-20 enhanced lin-42(n1089) mutant phenotypes including precocious alae formation. Consistent with these phenotypic results, we found that LIN-42A levels were decreased in kin-20(ok505) mutant worms. Like LIN-42, KIN-20 is important for both let-7 and lin-4 miRNA expression. Though it is possible the decrease in let-7 levels in kin-20 mutant worms is dependent on LIN-42A, our results more strongly suggest that KIN-20 impacts expression of the let-7 miRNA independently of LIN-42, since KIN-20 had no effect on a third constitutively-expressed, non-heterochronic miRNA target miR-58.1, and KIN-20 did not impact primary let-7 transcription. In addition, growth of lin-42(n1089) mutant worms on kin-20 RNAi caused a further reduction in let-7 levels. Altogether these results suggest that although KIN-20 regulates both LIN-42 and some miRNAs in C. elegans, KIN-20 mainly does so in a manner unpredicted by the inhibitory function of Doubletime (the KIN-20 homolog) on Period (the LIN-42 homolog) in Drosophila.
Materials and Methods
Nematode strains and culture conditions
The following C. elegans strains were used: wild type (WT) N2 Bristol, kin-20(ok505) (VC398), lin-42(ok2385) (RB1843), and lin-42(n1089) (MT2257). The integrated transgene wls79 contains ajm-1::gfp/MH27:: GFP and scm-1::GFP to allow visualization of seam cells. We crossed animals containing wls79 to VC398 to generate wls79;kin-20(ok505) worms. The integrated strain PQ462 contains 1568 bp of let-7 promoter sequence driving nuclear-localized GFP expression (plet-7B::GFP) (Kai et al. 2013). We crossed PQ462 to VC398 to generate plet-7B::GFP;kin-20(ok505).
Worms were maintained at 15° or 20° and synchronized by standard hypochlorite treatment. Starvation-arrested L1 worms were plated on E. coli OP50-seeded plates at 25° and collected at the appropriate time point. Larval stages correspond to the timing of development for WT N2 worms based on previously published time course analyses of worm development and molting at 25° (Jeon et al. 1999; Zisoulis et al. 2012), as lin-42 mutants develop somewhat asynchronously (Monsalve et al. 2011). We performed two-generation feeding RNAi experiments as described (Bracht et al. 2010) except that the IPTG concentration was increased to 10 mM. Briefly, L1 stage worms were grown on RNAi plates at 15°. Then synchronized, starved L1 progeny from these worms were grown on the same RNAi food at 25° until the desired stage before molecular or phenotypic analysis.
Brood counts were analyzed of synchronized, singled WT N2 or kin-20(ok505) worms grown at 15° or 25°. Parental worms were passaged to new plates over the course of the experiment to enable detection of all progeny. Death and dumpy phenotypes were analyzed in at least 500 adult animals grown on RNAi for two-generations as described above. The presence or absence of adult alae on the cuticle was measured in at least 20 synchronized animals after growth at 25° until the L4 or young adult stage. Alae was classified as complete if it extended continuously over all seam cells in three perfectly parallel ridges. Animals that had alae that was not complete and/or did not contain three perfectly parallel ridges were classified as having abnormal alae. Seam cell nuclei were counted in at least 19 synchronized animals grown at 25° until the L4 or young adult stage. Fluorescent microscopy analysis was performed on at least 25 synchronized animals after growth at 25° until the L3 or L4 stage. Fluorescent micrographs were captured under equivalent exposure times.
Statistical differences of sample phenotypes were analyzed as appropriate by Student’s t-tests or chi-square tests.
RNA analyses
Total RNA was extracted from synchronized, staged worm populations using TRIzol reagent (Life Technologies), and cDNA synthesis was completed with TaqMan microRNA assays (Thermo Fisher Scientific) or as previously described with random oligos or oligo dT (Van Wynsberghe et al. 2011a). qPCR was performed with TaqMan or SYBR Green reagents (Thermo Fisher Scientific) and 6.25 pmol of each primer (Table S1) on an ABI Prism 7900 or a Thermo Fisher Scientific QuantStudio 3 Real-time PCR system. Statistical differences of RNA levels between samples were analyzed by Student’s t-tests or two-way ANOVAs.
Protein analyses
Polyclonal antibodies against the C terminal sequence (KTSSSSSLLMLRDSQNE) of LIN-42 were raised in rabbits and purified by YenZym Antibodies, LLC. Western blotting was performed as described with this rabbit polyclonal antibody against LIN-42 (YenZym Antibodies, LLC) or a mouse monoclonal antibody against tubulin (Sigma-Aldrich) (Van Wynsberghe et al. 2011b). The HRP conjugated goat anti-rabbit or mouse secondary antibodies (ThermoFisher Scientific) were used and visualized on a ChemiDoc XRS+ (BioRad) system.
Data Availability
Reagents will be made available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6225893.
Results
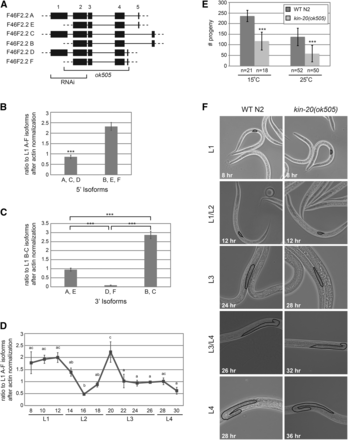
KIN-20 is a homolog of the Period protein kinase Doubletime in Drosophila and casein kinase I ε/δ in mammals (Banerjee et al. 2005). Like other circadian clock homologs in C. elegans, kin-20 is important for proper developmental timing (Banerjee et al. 2005). Previous research has utilized RNAi to ascertain the impact of kin-20 on developmental timing (Banerjee et al. 2005). To more completely abrogate kin-20 expression, we utilized the ok505 allele that causes an approximately 2.2 kb deletion in the kin-20 transcript that removes almost the entire kin-20 gene (Figure 1A). According to WormBase, the kin-20 gene encodes 6 distinct isoforms (labeled A-F) that each have different 5′ and 3′ UTRs of varying lengths and splice patterns. However, the internal sequence (exons 2-4) of all isoforms is shared (Figure 1A). In its simplest form there are 3 isoform pairs (A/E, C/B, and D/F) that share exons 2-5 (Figure 1A). In addition, one member of each pair (isoforms A, C and D) contains exon 1, while the other member (E, B, and F) initiates at an in frame AUG start codon at nt 6 of exon 2 (Figure 1A). By using primer sets that distinguish between KIN-20 isoforms that initiate at the 1st AUG and those that initiate at the 2nd AUG of KIN-20, we find that there is ∼3 fold more of the B, E and F isoforms than the A, C, and D isoforms at all time points throughout development (Figure 1B and Figure S1A). By using primer sets that distinguish between the differing 3′ isoforms, we find that there is ∼3 fold more of the B and C isoforms than the A and E isoforms, and ∼10 fold more of the A and E isoforms than the D and F isoforms at all time points throughout development (Figure 1C and Figure S1B). Thus, the B isoform is the major KIN-20 isoform expressed throughout development (Figure 1 and Figure S1). While the D and F isoforms are only weakly expressed, the A, E and C isoforms are all easily detectable throughout development (Figure 1 and Figure S1). In Drosophila Doubletime levels do not cycle (Kloss et al. 1998, 2001). By analyzing the expression pattern of all kin-20 isoforms throughout development, we find that kin-20 expression is relatively constant throughout development, except for the time prior to and surrounding the L2 molt (Figure 1D). This expression pattern is similar across all kin-20 isoforms (Figure S1).
KIN-20 is required for proper developmental timing. (A) Depiction of the kin-20 gene. According to WormBase there are 6 isoforms, A-F, each with alternative 5′ and 3′ UTRs. Each of the 6 isoforms is paired such that exons 2-5 are identical in each member of the pair. In addition, isoforms A, C and D contain exon 1, while isoforms E, B and F start at the 2nd in frame AUG start codon at nt 6 of exon 2. Untranslated sequences are depicted by dashed lines, and introns are depicted by solid lines. Regions impacted by RNAi or the ok505 allele are shown. (B-D) Synchronized, starved L1 WT N2 worms were grown at 25°C and collected every 2 hr from 8 to 30 hr after plating on food. RNA was extracted, reverse transcribed into cDNA, and analyzed by qPCR with primers located in exons 1 and 2 (to amplify isoforms A, C and D), in exons 2 and 3 (to amplify all isoforms A-F), or in exon 4 and each unique exon 5 (to amplify the distinct 3′ isoforms) of KIN-20. Data are shown from 3 independent experiments and was analyzed by a two-way ANOVA (***, P < 0.0005). Error bars show s.e.m. (B) The average ratios of the B, E and F isoforms and the A, C, and D isoforms are shown after normalization to the L1 A-F isoforms and actin. (C) The average ratios of the A, E isoforms, the D, F isoforms and the B, C isoforms are shown after normalization to the L1 B-C isoforms and actin. (D) The ratio of all isoforms of KIN20 after normalization to actin is shown. Letters indicate time points that are not significantly different from one another. More specifically, all time points marked “ac” are not significantly different from time points marked with an “a” or a “c”, but are significantly different from time points marked with only a “b”. (E) The average progeny number of kin-20(ok505) mutant worms compared to WT N2 worms was calculated from the indicated number of worms grown at 15°C and 25°C and analyzed by Student’s t-tests (***, P < 0.0005). Error bars show s.e.m. (F) Synchronized WT N2 worms were grown at 25°C until the times shown. Synchronized kin-20(ok505) worms were also grown at 25°C until the same stage in development as WT N2, as determined by gonad size. Representative images were taken at 400x magnification. Gonads are outlined.
The functional significance of these variations in kin-20 mRNA levels throughout development and between isoforms, as well as the protein expression patterns of KIN-20 are currently unclear. Despite the uncertainty surrounding these finer points, our experiments reveal that overall kin-20 is extremely important, as the fertility of C. elegans significantly drops ∼2 fold in kin-20(ok505) mutant worms relative to WT N2 worms when grown at either 15° or 25° (Figure 1E). As previously shown by RNAi (Banerjee et al. 2005), kin-20(ok505) mutant worms do not show any defects in the timing of alae formation (Figure S2A), but do exhibit precocious seam cell exit from the cell cycle during the late L4 stage (Figure S2B). In addition, the growth of C. elegans is dramatically slowed in kin-20(ok505) worms relative to WT N2 as measured by animal size and gonad development at 25° (Figure 1F and Figure S3). This growth delay became particularly apparent during the L3 stage, but may have initiated earlier in development (Figure 1F). In addition, the length of delay increased as the worms continued to develop (Figure 1F and Figure S3).
KIN-20 regulates LIN-42 expression
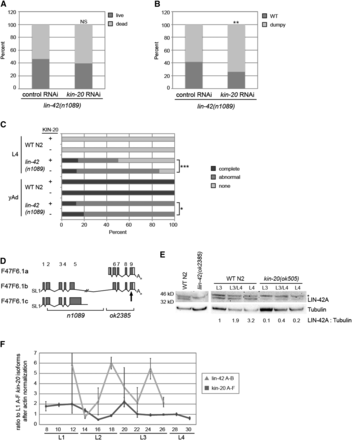
The KIN-20 homologs Doubletime and CK1ε/δ post-translationally regulate Period protein expression via phosphorylation, which marks Period for degradation (Price et al. 1998; Crane and Young 2014). LIN-42 is the Period protein homolog in C. elegans (Jeon et al. 1999). Thus, we asked if KIN-20 similarly impacts lin-42 expression in C. elegans by analyzing both lin-42 phenotypes and expression patterns in the presence and absence of kin-20. To do this we attempted to generate worms that were homozygous for both the lin-42(n1089) allele and the kin-20(ok505) allele. However, progeny of self fertilized C. elegans that were homozygous for the lin-42(n1089) mutation and heterozygous for the kin-20(ok505) mutation (or alternatively homozygous for the kin-20(ok505) mutation and heterozygous for the lin-42(n1089) mutation) yielded no progeny that were homozygous for both mutations (Table S2). Growth of lin-42(n1089) worms in the presence of kin-20 RNAi caused a slight, but non-significant increase in the proportion of worms that died by the young adult stage as compared to vector control RNAi (Figure 2A). Taken together, these observations suggest that when LIN-42 levels are reduced in the absence of KIN-20, as is seen in the double mutant, but not through RNAi treatment, lethality results.
KIN-20 promotes LIN-42 expression. (A-C) lin-42(n1089) mutant worms were subjected to vector control RNAi or kin-20 RNAi and analyzed after growth at 25°C. The percentage of dead worms (A) and dumpy worms (B) after growth for 2 days are shown (N > 150), and were analyzed by a chi-square test (***, P < 0.0005). NS equals non-significant. (C) The presence of alae was analyzed in late L4 and yAd worms after growth on vector control or kin-20 RNAi. N > 20. Alae was classified as complete if it extended continuously over all seam cells in three parallel ridges. Alae was classified as abnormal if it was not complete and/or did not contain three perfectly parallel ridges. (D) Depiction of the lin-42 gene, based on WormBase. The n1089 and ok2385 alleles are marked below the gene diagrams. The site targeted by the LIN-42 antibody is marked with an arrow. (E) Protein was extracted from mixed stage WT N2 and lin-42(ok2385) mutant worms, and synchronized WT N2 worms and kin-20(ok505) mutant worms and analyzed by western blotting for LIN-42 and tubulin. Asterisk denotes non-specific bands. A representative blot from 3 independent experiments is shown. The ratio of LIN-42 A to tubulin after normalization to LIN-42A in WT N2 at the L3 stage is shown. (F) Synchronized WT N2 worms were grown at 25°C and collected every 2 hr as described in Fig. 1 before analysis by qPCR for all kin-20 isoforms (A-F) or the A and B isoforms of lin-42. The ratio to all isoforms of KIN20 at L1 after normalization to actin is shown. Data are shown from 3 independent experiments. Error bars show s.e.m.
We also found that lin-42 mutant phenotypes were enhanced in the absence of kin-20. Due to molting defects lin-42 mutant worms exhibit a dumpy phenotype as adults (Monsalve et al. 2011). We find that significantly more lin-42(n1089) mutant worms display this dumpy phenotype when subjected to kin-20 RNAi as compared to vector control RNAi (Figure 2B). As a member of the heterochronic pathway, lin-42 mutant worms have previously been shown to exhibit a precocious alae phenotype (Abrahante et al. 1998; Van Wynsberghe and Pasquinelli 2014). Our analysis of alae formation found that many lin-42(n1089) worms exhibited abnormal alae formation, as defined by alae that was either incomplete (did not extend extend continuously over all seam cells) or was not in three perfectly parallel ridges. At the young adult stage, significantly more animals exhibit complete alae in lin-42(n1089) worms grown on kin-20 RNAi compared to vector control RNAi (Figure 2C). In addition, at the L4 stage, though a similar proportion of lin-42(n1089) worms have complete alae, significantly more lin-42(n1089) worms exhibit precocious alae formation (as seen by the presence of either complete or abnormal alae) when grown on kin-20 RNAi compared to vector control RNAi (Figure 2C).
LIN-42 has 3 isoforms (Figure 2D) (Edelman et al. 2016). The largest isoform, B, contains sequence found in both the A and C isoforms (Figure 2D). Work in other labs has suggested that the LIN-42A and B isoforms are of greater functional importance than LIN-42C (Tennessen et al. 2006; Monsalve et al. 2011; Edelman et al. 2016). Using an antibody developed against the C terminal region of LIN-42, we were able to clearly visualize the LIN-42A isoform (Figure 2E, Figure S4). In the absence of KIN-20 we found that LIN-42A levels are severely decreased (Figure 2E, Figure S4). To further analyze the effect of kin-20 on lin-42 expression, we compared the expression patterns of lin-42 isoforms A and B to kin-20 expression throughout all four larval stages in WT N2 worms (Figure 2F). As expected, lin-42 levels oscillate throughout development (Figure 2F). The significant decrease in kin-20 mRNA levels found in the L2 stage is followed by an increase in lin-42 mRNA levels, while the significant increase in kin-20 mRNA levels found in the early L3 stage is followed by a decrease in lin-42 mRNA levels (Figure 2F). Altogether, our work provides a causal link for our observations that kin-20 knockdown enhances lin-42 mutant phenotypes by further reducing LIN-42 levels. This suggests that unlike its homologs KIN-20 promotes LIN-42 expression.
KIN-20 regulates expression of some microRNAs
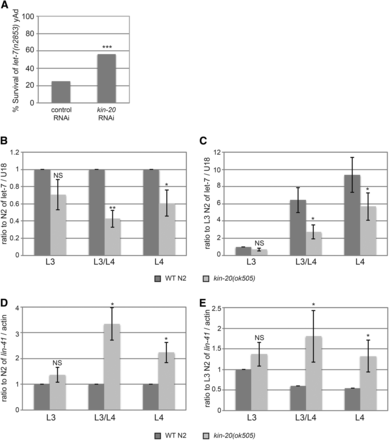
We and others have previously shown that LIN-42 is a transcription factor that acts to inhibit transcription of a multitude of genes including small RNAs called microRNAs (Monsalve et al. 2011; Mcculloch and Rougvie 2014; Van Wynsberghe et al. 2014). Thus, we hypothesized that KIN-20 might also regulate microRNAs and other genes through its effects on LIN-42. Indeed, Banerjee et al. have previously shown that kin-20 RNAi can rescue the lethal bursting phenotype that occurs in worms with a mutation in the let-7 microRNA (Banerjee et al. 2005). In agreement with these previous findings, we demonstrated that RNAi against kin-20 significantly rescues the lethal bursting phenotype seen in let-7(n2853) mutant worms after the L4 molt when grown at 25° (Figure 3A), though not as completely as previously reported (Banerjee et al. 2005). Since RNAi against lin-42 or a loss-of-function lin-42 mutation also rescues the let-7 bursting phenotype by increasing let-7 levels (Banerjee et al. 2005; Van Wynsberghe et al. 2014; Perales et al. 2014), we next asked if the ability of kin-20 RNAi to rescue this let-7 phenotype was also due to alterations in let-7 levels. We analyzed mature let-7 levels at the third larval stage (L3), the L3 molt (L3/L4) and the fourth larval stage (L4) at 25° according to size and gonad development (Figure 1F and Figure S3) in kin-20(ok505) and WT N2 worms (Figure 3B-C). We found that let-7 levels decreased at all time points and that this decrease was significant at the L3 molt and L4 time points (Figure 3B-C). This decrease in let-7 coincided with a significant ∼2-3 fold increase in mRNA levels of the let-7 target gene, lin-41, in kin-20(ok505) mutant worms relative to WT N2 worms (Figure 3D-E).
KIN-20 is important for let-7 miRNA expression. (A) The percentage of worms alive after growth for 2 days at 25°C on vector control RNAi or kin-20 RNAi is shown (N > 200), and was analyzed by a chi-square test (***, P < 0.0005). (B-E) Synchronized WT N2 or kin-20(ok505) mutant worms were grown at 25°C and collected at the L3, L3/L4, or L4 stage based on size and gonad development. RNA was extracted and analyzed by qPCR after reverse transcription for let-7 (B-C) or lin-41 (D-E). The average and s.e.m. of RNA levels from at least 6 independent experiments after normalization to U18 mRNA (B-C) or actin mRNA (D-E) are graphed and were analyzed by Student’s t-tests (*, P < 0.05; **, P < 0.005). NS equals non-significant. Levels are shown relative to WT N2 at all time points (B,D) or specifically to WT N2 at the L3 stage (C,E).
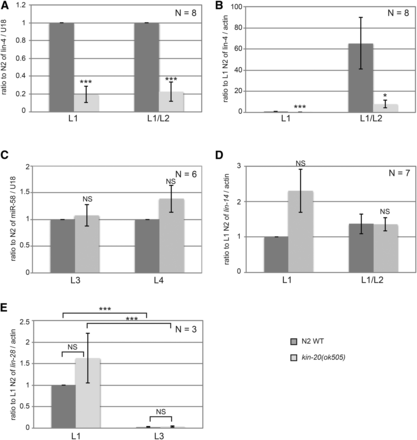
Some regulators of miRNA biogenesis act specifically on a single miRNA, while others, like LIN-42, act more globally to affect biogenesis of multiple miRNAs (Finnegan and Pasquinelli 2013). To determine if kin-20 affected other miRNAs, we analyzed the effect of kin-20 on two other miRNAs: lin-4 and miR-58.1. lin-4 is also a member of the heterochronic pathway and is first expressed in the mid-L1 stage (Bracht et al. 2010), while miR-58.1 is non-heterochronic gene that is expressed throughout development (Abbott 2011). Like let-7, lin-4 miRNA levels were significantly decreased in kin-20(ok505) relative to WT N2 worms (Figure 4A-B). In contrast, levels of miR-58.1 were not altered in kin-20(ok505) relative to WT N2 worms at L3 and L4 stages (Figure 4C). lin-4 functions early in the heterochronic pathway by regulating both lin-14 and lin-28 (Ambros 1989). Consistent with a decrease in lin-4, we find that lin-14 mRNA levels are increased early in development in kin-20(ok505) relative to WT N2 worms (P = 0.076), though there is no effect on lin-14 mRNA at a later stage (Figure 4D). We also find that lin-28 mRNA levels are increased, though not significantly, in kin-20(ok505) relative to WT N2 worms at the L1 stage (Figure 4E). However, given the importance of lin-4 in inhibiting lin-28 mRNA levels (Ambros 1989), and our finding that lin-4 levels are significantly reduced in kin-20(ok505) worms (Figure 1A-B), it is surprising that lin-28 mRNA levels are significantly decreased in the L3 stage (Figure 4E). This normal expression pattern of lin-28 mRNA suggests that though KIN-20 may in part regulate let-7 via its effects on lin-4, KIN-20 likely predominantly regulates let-7 downstream of its impacts on lin-4.
Effect of KIN-20 on other microRNAs and heterochronic genes. Total RNA was extracted from synchronized WT N2 and kin-20(ok505) mutant worms at the time points shown, and analyzed as in Fig. 3 for lin-4 (A-B), miR-58.1 (C), or lin-4 targets lin-14 (D) and lin-28 (E). The average and s.e.m. of RNA levels from multiple independent experiments after normalization to U18 (A-C) or actin mRNA (D-E) are shown and were analyzed by Student’s t-tests (*, P < 0.05; ***, P < 0.0005). NS equals non-significant.
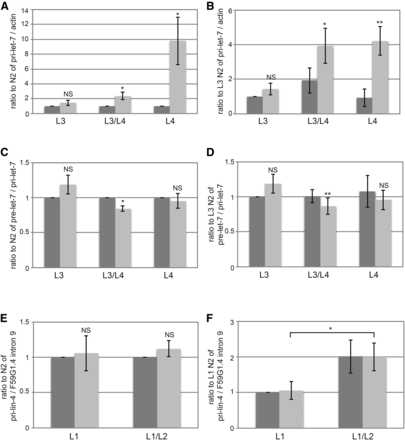
Decreased levels of mature let-7 could be due to a decrease in transcription of the let-7 gene, decreased processing of primary or precursor let-7 RNAs, or decreased stability of mature let-7. To distinguish among these possibilities, we analyzed the levels of these RNAs involved in let-7 biogenesis in the same samples for which mature let-7 levels had also been analyzed. Transcription initiation at two distinct sites yields two primary let-7 transcripts that are both subsequently spliced to yield a third primary let-7 transcript that is ultimately processed into precursor let-7 (Bracht et al. 2004; Van Wynsberghe et al. 2011b). Levels of all three primary let-7 transcripts were analyzed with a single primer set. Primary let-7 levels were unchanged at the L3 stage, but were significantly increased later in development in kin-20(ok505) mutant worms relative to WT (Figure 5A-B). Levels of precursor let-7 varied slightly at each stage tested in kin-20(ok505) mutant worms relative to WT, but overall suggested that pre-let-7 levels were either minimally altered or that any impacts on pre-let-7 were negated by its rapid processing into mature let-7 (Figure 5C-D).
Effect of KIN-20 on microRNA biogenesis. Total RNA samples from synchronized WT N2 and kin-20(ok505) mutant worms that were analyzed for mature let-7 and mature lin-4 in Figs. 3 and 4 respectively were also analyzed for pri-let-7 (A-B), pre-let-7 (C-D), and pri-lin-4 (E-F). The average and s.e.m. of RNA levels from 6 (A-D) or 7 (E-F) independent experiments after normalization to actin mRNA (A-B), pri-let-7 levels (C-D), or F59G1.4 intron 9 are shown and were analyzed by Student’s t-tests (*, P < 0.05; **, P < 0.005). NS equals non-significant. Levels are shown relative to WT N2 at all time points (A,C,E) or specifically to WT N2 at the L3 stage (B,D) or L1 stage (F).
We also analyzed the effect of KIN-20 on primary lin-4 in the same samples for which mature lin-4 had also been analyzed. Primary lin-4 transcription initiates from two distinct start sites encoded in the 9th intron of the ubiquitously expressed F59G1.4 gene (Bracht et al. 2010). Levels of both primary lin-4 transcripts were analyzed with a single primer set and normalized to F59G1.4 intronic sequence. Unlike primary let-7, we found that kin-20 had no impact on primary lin-4 levels at either time point (Figure 5E-F), suggesting that KIN-20 acts post-transcriptionally to regulate lin-4 expression.
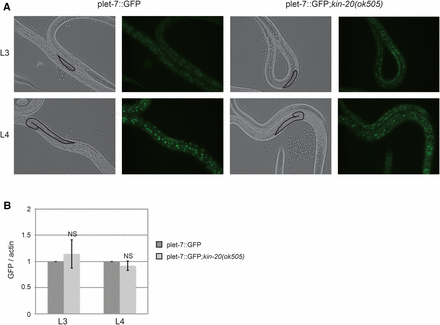
To assess if KIN-20 acted as a transcriptional or post-transcriptional regulator of pri-let-7, we utilized an integrated reporter that expresses GFP from ∼1568 bp of let-7 promoter sequence (Kai et al. 2013), and visualized let-7 expression in the seam cells. As expected (Kai et al. 2012), let-7 promoter driven expression of GFP was stronger at the L4 stage compared to the L3 stage (Figure 6A). However, there was no consistent difference in GFP expression in the presence vs. absence of kin-20 as visualized by fluorescent microscopy (Figure 6A). Because this is a stable GFP reporter, we also analyzed GFP mRNA levels to better understand the impact of kin-20 on let-7 transcription. Quantitative real time PCR analysis showed that GFP mRNA levels were unchanged in kin-20(ok505) mutant worms relative to WT N2 worms at both the L3 and L4 stages (Figure 6B). Altogether these results suggest that in the absence of kin-20 a blockage of primary let-7 processing causes an increase in pri-let-7 levels coincident with a decrease in mature let-7.
KIN-20 does not impact let-7 transcription. (A) Representative images of WT or kin-20(ok505) mutant worms expressing the plet-7::GFP reporter at L3 and L4 stages. Fluorescent micrographs were captured under equivalent exposure times. (B) Total RNA was isolated from synchronized WT N2 or kin-20(ok505) mutant worms expressing the plet-7::GFP reporter during the L3 and L4 stages. The level of GFP after actin mRNA normalization relative to WT N2 worms was calculated from 3 independent experiments. Error bars show s.e.m. NS equals non-significant.
KIN-20 mainly regulates let-7 independently of its effects on LIN-42
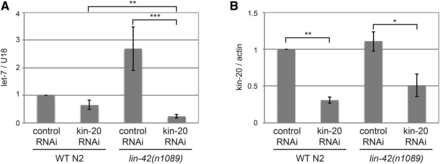
To further test if KIN-20 affected let-7 miRNA expression by regulating LIN-42 protein levels, we analyzed the levels of let-7 miRNA in WT N2 or lin-42(n1089) worms grown in the presence of vector control or kin-20 RNAi (Figure 7). RNAi against kin-20 did not cause the growth delay seen in kin-20(ok505) mutant worms (Figure S5). As expected, let-7 levels of worms grown on vector control RNAi were ∼2.5 fold increased in lin-42(n1089) mutant worms relative to WT N2 worms (Figure 7A) (Van Wynsberghe et al. 2014). Levels of let-7 were only slightly decreased in WT N2 worms treated with kin-20 RNAi (Figure 7A). This smaller effect, as compared to the change in let-7 levels in a kin-20 mutant (Figure 3), was likely due to the only ∼2-3 fold decrease in kin-20 levels from kin-20 RNAi treatment (Figure 7B). Despite this, knockdown of kin-20 levels by RNAi significantly reduced let-7 levels by 10 fold in lin-42(n1089) mutant worms (Figure 7A). Levels of let-7 were also significantly decreased by ∼3 fold in lin-42(n1089) mutant worms treated with kin-20 RNAi compared to WT N2 worms treated with kin-20 RNAi (Figure 7A). Because let-7 levels are significantly different in the absence of both lin-42 and kin-20 compared to just the absence of lin-42 or kin-20, this together with our other data suggests that KIN-20 predominantly acts independently of its effects on LIN-42 to regulate let-7 levels.
KIN-20 mainly impacts miRNA levels independently of LIN-42. Total RNA was extracted from synchronized WT N2 or lin-42(n1089) mutant worms subjected to 2-generation vector control RNAi or kin-20 RNAi and collected at the L3/L4 stage. RNA was analyzed by qPCR after reverse transcription. The average and s.e.m. of RNA levels from 3 independent experiments are shown and were analyzed by Student’s t-tests (*, P < 0.05; **, P < 0.005; ***, P < 0.0005). (A) Levels of let-7 miRNA after normalization to U18 mRNA relative to WT N2 subjected to vector control RNAi. (B) Levels of kin-20 mRNA after actin mRNA normalization relative to WT N2 subjected to vector control RNAi.
Discussion
Here we further characterize the Period protein kinase homolog KIN-20 and demonstrate that KIN-20 regulates both the Period protein homolog LIN-42 and specific miRNAs like let-7 and lin-4. We show that though all KIN-20 isoforms are expressed throughout development, there is great variation in the levels of expression of these isoforms and that the B isoform of kin-20 is the most expressed (Figure 1 and Figure S1). We find that kin-20 mutant worms have decreased progeny numbers, grow slowly and exhibit aberrant seam cell development, but not alae production (Figure 1 and Supplementary Figures 2 and 3). We show that LIN-42A levels are decreased and lin-42 mutant phenotypes are enhanced when KIN-20 levels are decreased (Figure 2). In addition, in the absence of KIN-20, mature let-7 levels decrease concordantly with an increase in primary-let-7 levels (Figures 3 and 5). KIN-20 similarly affects mature lin-4 levels, but not mature miR-58.1 levels, and pri-lin-4 levels are not altered in kin-20 mutant worms (Figures 4 and 5). KIN-20 mediates these effects on let-7 and lin-4 post-transcriptionally because GFP mRNA and protein levels, when placed under the control of the let-7 promoter, are not altered in the absence of KIN-20 (Figure 6). KIN-20 impacts LIN-42 and let-7, and LIN-42 has been previously shown to inhibit let-7 expression (Mcculloch and Rougvie 2014; Van Wynsberghe et al. 2014; Perales et al. 2014). However, because let-7 levels significantly differ in lin-42 mutant worms that express decreased levels of kin-20, we can conclude that KIN-20 mainly regulates let-7 independently of LIN-42. These results uncover a new mechanism used to control both the conserved Period protein homolog LIN-42 and the important, conserved microRNA let-7, and thus developmental and rhythmic processes.
Developmental timing in C. elegans is maintained by the heterochronic pathway, which is comprised of a complex network of genes (Rougvie and Moss 2013). When absent, heterochronic genes either cause precocious developmental phenotypes or reiteration of, and thus retarded, developmental phenotypes. Gain-of-function mutations in heterochronic genes cause the opposite developmental effect. Measurement of developmental delays in C. elegans is most commonly done through analysis of hypodermal seam cells, which exhibit regular division patterns throughout larval growth before fusing and secreting alae, a series of cuticular ridges, in the young adult stage (Resnick et al. 2010). Consistent with previously reported results (Banerjee et al. 2005), we find that kin-20(ok505) mutant worms precociously exit the cell cycle at late L4, represented by a decrease in the number of hypodermal seam cell nuclei at this stage (Figure S2B). Additionally, kin-20(ok505) mutant worms exhibit wild type timing of alae formation (Figure S2A) (Banerjee et al. 2005). However, kin-20 RNAi enhances the precocious alae phenotype in lin-42(n1089) mutant worms (Figure 2C). In addition, kin-20(ok505) mutants exhibit altered expression of multiple heterochronic genes including lin-42, let-7 and lin-4 (Figures 2-4). Despite these impacts on gene expression, the finding that KIN-20 itself does not exhibit aberrant alae development, and therefore the altered developmental timing typical of heterochronic genes, suggests that KIN-20 is not a traditional heterochronic gene.
Like other Period proteins, expression of LIN-42, the C. elegans Period protein homolog, oscillates throughout development (Jeon et al. 1999; Tennessen et al. 2006; Monsalve et al. 2011; Edelman et al. 2016). Given the role of Doubletime in regulating Period, we hypothesized that the Doubletime homolog KIN-20 would similarly impact LIN-42. However, we found that levels of the LIN-42A isoform were decreased and that multiple lin-42 mutant phenotypes were enhanced in kin-20(ok505) mutant worms (Figure 2). Since LIN-42A levels were decreased in kin-20(ok505), we might expect kin-20(ok505) worms to phenocopy lin-42(ok2385) worms, which delete LIN-42A and contain a large deletion in the C-terminal region of LIN-42B (Edelman et al. 2016). Like lin-42(ok2385), kin-20(ok505) worms have a significantly reduced brood size, growth delays, and precocious alae (Figures 1 and 2) (Edelman et al. 2016). Similarly, lin-42(n1089) mutant worms, which contain a large deletion at the N terminus of lin-42, when grown in the absence of kin-20 might be expected to phenocopy lin-42(ox461) worms, which lack all lin-42 isoforms (Edelman et al. 2016). Unfortunately, we were unable to analyze the phenotypes of lin-42(n1089);kin-20(ok505) due to their lethality (Table S2). However, we did analyze the phenotypes of lin-42(n1089) worms subjected to kin-20 RNAi. Despite kin-20 RNAi only decreasing kin-20 levels ∼2-3 fold (Figure 7B), we found that lin-42 mutant phenotypes were enhanced after kin-20 RNAi treatment (Figure 2). lin-42(ox461) worms exhibited enhanced larval arrest and growth delays (Edelman et al. 2016). In contrast, lin-42(n1089) worms subjected to kin-20 RNAi did not show any significant growth delays (Figure S5). Though the proportion of worms exhibiting some type of precocious alae (partial or complete) did not change in lin-42(ox461) worms, more lin-42(ox461) worms exhibited complete alae (Edelman et al. 2016). We also found synergistic effects of lin-42 and kin-20 on alae production (Figure 2C). However, we found that the proportion of worms exhibiting some type of precocious alae (abnormal or complete), but not the number of worms exhibiting complete alae, significantly increased in lin-42(n1089) worms subjected to kin-20 RNAi (Figure 2C). Thus, our results suggest that both lin-42 and kin-20 act in similar pathways, in addition to their distinct, crucial functions in development. Altogether, these results suggest that instead of destabilizing LIN-42, KIN-20 acts to stabilize or promote LIN-42A expression.
There are three LIN-42 isoforms that have each been shown to be important for proper developmental timing (Edelman et al. 2016). However, there is still much to be determined about whether the isoforms have distinct functions and/or expression patterns. LIN-42C and the N terminal region of LIN-42B contain the conserved protein interaction (PAS) domain characteristic of Period proteins, while LIN-42A and the C terminal region of LIN-42B contain the conserved nuclear localization signal and the SYQ and LT regions which contain multiple ser, tyr and gln or leu and thr amino acids respectively (Figure 2D) (Jeon et al. 1999; Tennessen et al. 2006). LIN-42A and the C terminal region of LIN-42B are thought to contain the most important regions of LIN-42 since mutations in LIN-42C and the N terminus of LIN-42B can be rescued by overexpression of LIN-42A (Tennessen et al. 2006). In contrast, overexpression of LIN-42C does not rescue mutations in LIN-42A or the C terminus of LIN-42B (Tennessen et al. 2006). In addition, LIN-42A is particularly important for regulating molting and seam cell development in C. elegans (Monsalve et al. 2011). Unfortunately, our western blotting analysis only enabled detection of the LIN-42A isoform. Thus it is still unclear if and how KIN-20 regulates the other isoforms of LIN-42. To start to address this issue we compared levels of lin-42 A and B mRNA to levels of all kin-20 isoforms by qRT-PCR (Figure 2F). In Drosophila, Period engages in an autoregulatory negative feedback loop to inhibit transcription of the Period gene (Peschel and Helfrich-Förster 2011). Thus a decrease in kin-20 levels, as seen in the L2 stage (Figure 2F), would cause a decrease in LIN-42A levels followed by a subsequent increase in lin-42 mRNA levels. Similarly, an increase in kin-20 levels, as seen at the beginning of L3 (Figure 2F), would cause an increase in LIN-42A levels, and thus a subsequent decrease in lin-42 mRNA. Though both of these associations do occur (Figure 2F), more studies are needed to clearly show the impact of KIN-20 on the expression of individual LIN-42 isoforms. In addition, it is unclear if such an autoregulatory negative feedback loop even exists in C. elegans, since previous work in the Rougvie lab has shown that lin-42 levels still oscillate in the absence of functional LIN-42 protein (Jeon et al. 1999). Regardless, because we find that lin-42 mutant phenotypes are enhanced in kin-20(ok505) mutant worms, it is most likely that if KIN-20 regulates the other LIN-42 isoforms it does so in a similar manner to its effects on LIN-42A. Based on its homology with Doubletime, it is also most likely that KIN-20 mediates these effects via phosphorylation.
LIN-42 has previously been shown to act as a transcription factor that negatively regulates the expression of numerous target genes including the miRNAs let-7, lin-4 and miR-58.1 (Mcculloch and Rougvie 2014; Van Wynsberghe et al. 2014; Perales et al. 2014). Thus, because of its impact on LIN-42, we hypothesized that KIN-20 might also regulate these miRNAs. This hypothesis was further supported by the finding that kin-20 RNAi rescued the lethal let-7(n2853) bursting phenotype (Figure 3A) (Banerjee et al. 2005), though we found that the degree of rescue was less than previously reported (Banerjee et al. 2005). The n2853 allele is a temperature-sensitive point mutation in the seed sequence of the mature miRNA that decreases the levels of mature let-7 by more than 5 fold relative to WT (Reinhart et al. 2000; Bagga et al. 2005; Chatterjee and Großhans 2009; Zisoulis et al. 2012). Reduced let-7 levels then cause bursting through the vulva after the L4 molt at 25° (Reinhart et al. 2000). When their levels are reduced, various members of the heterochronic pathway and other pathways have been found to rescue this lethal phenotype. For example, lin-42 RNAi rescues the lethal let-7(n2853) bursting phenotype by causing an ∼2.5 fold increase in let-7 levels (Banerjee et al. 2005; Van Wynsberghe et al. 2014). Surprisingly, we found that instead of expressing increased let-7 levels, kin-20(ok505) mutant worms showed ∼2 fold decreased levels of let-7 relative to WT (Figure 3B-C) and a corresponding increase in the let-7 target lin-41 (Figure 3D-E). This further reduction in let-7 levels would be expected to increase let-7(n2853) lethality, not reduce it as we found (Figure 3A). Thus, our results suggest that KIN-20 suppresses the let-7 bursting phenotype in a manner that is independent of the heterochronic pathway. For example, KIN-20 may impact the expression of other genes involved in vulva formation in order to rescue this lethal phenotype.
KIN-20 impacts miRNA expression differently from LIN-42 since levels of the lin-4 miRNA, but not the constitutively expressed miR-58.1 miRNA were decreased in kin-20(ok505) mutant worms (Figure 4A-C). lin-4 acts early in the heterochronic pathway to regulate developmental timing (Ambros 1989). Surprisingly, levels of the lin-4 targets lin-14 and lin-28 were only increased initially in kin-20(ok505) mutant worms (Figure 4D-E). LIN-28 normally acts to downregulate expression of mature let-7, and let-7 levels only increase during the L3 stage after LIN-28 expression decreases as a result of lin-4 expression (Resnick et al. 2010; Lee et al. 2016). Thus, it is possible that KIN-20 could regulate let-7 indirectly via lin-4 and lin-28. However, the finding that lin-28 mRNA levels decrease normally in the L3 stage suggests that if KIN-20 regulates let-7 via lin-4 and lin-28, KIN-20 must also regulate let-7 downstream of its effects on lin-4 (Figure 4E).
Unlike LIN-42, we find that KIN-20 acts post-transcriptionally to regulate levels of mature let-7 and lin-4. KIN-20 had no effect on primary lin-4 levels (Figure 5E-F). Though primary let-7 levels were increased in kin-20(ok505) mutant worms (Figure 5A-B), transcription from the let-7 promoter was not affected since GFP mRNA levels were unchanged in kin-20(ok505) mutant worms (Figure 6). The increase in primary let-7 levels concordant with the decrease in mature let-7 levels suggests that KIN-20 does not impact mature let-7 stability. In addition, the fact that precursor let-7 levels remain mostly unchanged (Figure 5C-D) suggests that KIN-20 most likely regulates the processing of primary let-7 into precursor let-7. There are many proteins that regulate mature let-7 production at all steps in miRNA biogenesis (Finnegan and Pasquinelli 2013; Lee et al. 2016). Although the exact mechanism that KIN-20 utilizes to have this effect is still unclear, KIN-20 likely impacts another let-7 regulator through phosphorylation.
Our results suggest that KIN-20 positively regulates both LIN-42 and let-7. Previous work in our lab and others has shown that LIN-42 also negatively regulates let-7 (Mcculloch and Rougvie 2014; Van Wynsberghe et al. 2014; Perales et al. 2014). Thus, though it is possible that KIN-20 positively regulates LIN-42A to positively regulate let-7 expression, this model does not fit with previously published reports that LIN-42 acts as an inhibitor of miRNA transcription (Mcculloch and Rougvie 2014; Van Wynsberghe et al. 2014; Perales et al. 2014). Though we cannot rule out that KIN-20 regulates let-7 through its effects on LIN-42A, our data suggests that instead KIN-20 mainly regulates let-7 independently of its effects on LIN-42. First, LIN-42A levels decrease in the absence of kin-20 (Figure 2). Because lin-42(n1089) specifically eliminates LIN-42B and C isoforms, reducing kin-20 levels by RNAi in lin-42(n1089) worms will reduce LIN-42A levels and thus should enhance lin-42 knock-out phenotypes. Accordingly, we find that lin-42(n1089) worms subjected to kin-20 RNAi do indeed exhibit enhanced lin-42 mutant phenotypes (Figure 2). Since LIN-42 represses let-7 expression (Mcculloch and Rougvie 2014; Van Wynsberghe et al. 2014; Perales et al. 2014), and we have found that LIN-42A levels decrease in the absence of kin-20 (Figure 2), we would expect let-7 levels to increase in a kin-20 mutant that expresses decreased LIN-42A levels. However, the opposite occurs, suggesting that kin-20 must impact let-7 expression independently of its affects on LIN-42 (Figure 3). Second, LIN-42 regulates ∼30% of all miRNAs at the L4 stage (Van Wynsberghe et al. 2014; Perales et al. 2014). Thus, if KIN-20 acted predominantly through LIN-42, we would expect KIN-20 to similarly affect multiple miRNAs. Instead, we find that KIN-20 does not affect miR-58.1 (Figure 5), which is regulated by LIN-42 (Van Wynsberghe et al. 2014). In addition, since LIN-42 transcriptionally regulates let-7 (Mcculloch and Rougvie 2014; Van Wynsberghe et al. 2014; Perales et al. 2014), we would expect KIN-20 to also act at the transcriptional level if it mainly impacted let-7 expression via LIN-42. However, our results suggest that KIN-20 regulates let-7 post-transcriptionally (Figure 6). To further test if KIN-20 regulates let-7 via LIN-42, we analyzed let-7 levels in lin-42(n1089) mutant worms treated with kin-20 RNAi. If KIN-20 acted solely via LIN-42, we would expect let-7 levels to be the same in lin-42(n1089) worms treated with vector control or kin-20 RNAi. Instead we find that let-7 levels are significantly reduced in lin-42(n1089) worms treated with kin-20 RNAi compared to either lin-42(n1089) worms treated with vector control RNAi or WT N2 worms treated with kin-20 RNAi (Figure 7). Thus the decrease in both lin-42 and kin-20 levels acts to enhance the reduction in let-7 levels. In summary, though these results do not exclude the possibility that the decrease in let-7 levels in kin-20 mutant worms is dependent on LIN-42A, these results strongly support another mechanism, that is independent of LIN-42A, by which kin-20 regulates let-7 levels.
Altogether these results identify KIN-20 as a new, important regulator of LIN-42 and the conserved lin-4 and let-7 miRNAs. These results also highlight several important differences between KIN-20 and its homologs Doubletime and CKIε/δ, and increase our understanding of how rhythmic and developmental processes are regulated in C. elegans.
Acknowledgments
We thank members of the Van Wynsberghe laboratory and Dr. Amy E. Pasquinelli for their suggestions and critical reading of this manuscript. We thank members of the Van Wynsberghe lab including Nora Landells, Shannon Lacy, Carolyn Robb, and McKenzie Wallace for their work related to this project. We thank the Caenorhabditis Genetics Center for worm strains. This work was funded by Colgate University (P.M. V.W.).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6225893.
Communicating editor: J. Kim