-
PDF
- Split View
-
Views
-
Cite
Cite
Lizziane K Winkelströter, Stephen K Dolan, Thaila Fernanda dos Reis, Vinícius Leite Pedro Bom, Patrícia Alves de Castro, Daisuke Hagiwara, Raneem Alowni, Gary W Jones, Sean Doyle, Neil Andrew Brown, Gustavo H Goldman, Systematic Global Analysis of Genes Encoding Protein Phosphatases in Aspergillus fumigatus, G3 Genes|Genomes|Genetics, Volume 5, Issue 7, 1 July 2015, Pages 1525–1539, https://doi.org/10.1534/g3.115.016766
Close - Share Icon Share
Abstract
Aspergillus fumigatus is a fungal pathogen that causes several invasive and noninvasive diseases named aspergillosis. This disease is generally regarded as multifactorial, considering that several pathogenicity determinants are present during the establishment of this illness. It is necessary to obtain an increased knowledge of how, and which, A. fumigatus signal transduction pathways are engaged in the regulation of these processes. Protein phosphatases are essential to several signal transduction pathways. We identified 32 phosphatase catalytic subunit-encoding genes in A. fumigatus, of which we were able to construct 24 viable deletion mutants. The role of nine phosphatase mutants in the HOG (high osmolarity glycerol response) pathway was evaluated by measuring phosphorylation of the p38 MAPK (SakA) and expression of osmo-dependent genes. We were also able to identify 11 phosphatases involved in iron assimilation, six that are related to gliotoxin resistance, and three implicated in gliotoxin production. These results present the creation of a fundamental resource for the study of signaling in A. fumigatus and its implications in the regulation of pathogenicity determinants and virulence in this important pathogen.
Aspergillus fumigatus is a filamentous fungus that is able to live in the soil and is capable of causing a wide variety of noninvasive and invasive diseases in mammalian hosts, termed aspergillosis (Greenberger 2002; Dagenais and Keller 2009). One of these human diseases, invasive aspergillosis (IA), has a high frequency of mortality in immunocompromised patients. Aspergillosis is considered a multifactorial disease, because several pathogenicity determinants are required for the establishment of infection. The main factors are hypoxia stress resistance, iron assimilation, gliotoxin production (depending on the immune status of the host), and thermophily (Wezensky and Cramer 2011; Schrettl and Haas 2011; Hartmann et al. 2011; Carberry et al. 2012; Grahl et al. 2012; Scharf et al. 2012; Moore 2013; Ding et al. 2014; Chotirmall et al. 2014; Haas 2014). To effectively combat this life-threatening disease, it is essential to identify and understand cellular mechanisms of how these pathogenicity determinants are coordinated and which signaling molecules are essential for these virulence programs.
Protein kinases and phosphatases are responsible for regulating the continuous equilibrium between protein phosphorylation and dephosphorylation states. The addition or subtraction of phosphate residues by the respective enzymes occurs at specific amino acids such as serine, threonine, and tyrosine residues. Nucleophilic attack of the phosphate ester moiety is the main mechanism of phosphate dephosphorylation (Sanvoisin and Gani 2001; Williams 2004). There are two main families of phosphatases: the serine/threonine (S/T) protein phosphatases and the protein tyrosine phosphatases (PTP) (Pao et al. 2007; Shi 2009). S/T phosphatases are made of three subfamilies: phosphoprotein phosphatases (PPPs), metal-dependent protein phosphatases (PPMs), and aspartate-based protein phosphatases, comprising the transcription factor IIF–interacting C-terminal domain phosphatase (FCP/SCP) and haloacid dehalogenase (HAD) classes (Shi 2009; Zhang et al. 2010). PTPs are classified into classical protein-tyrosine phosphatases (PTPs), dual-specificity phosphatases (DSPs), low-molecular-weight phosphatases (LMW-PTP), and the CDC25 class phosphatases (Andersen et al. 2001; Pao et al. 2007; Moorhead et al. 2007).
Filamentous fungal phosphatases have been characterized in more detail in A. nidulans and Neurospora crassa (Son and Osmani 2009; Ghosh et al. 2014). In A. nidulans, 28 protein phosphatase catalytic subunit genes were identified and systematic deletion analysis identified four essential phosphatases and four required for normal growth (Son and Osmani 2009). The authors have identified several phosphatases involved in different aspects of cell division and mitosis. However, they have not performed an extensive phenotypic analysis of these mutants. More recently, Brown et al. (2013) have shown seven of these phosphatases as being involved in cellulase (and in some cases also hemicellulase) production in A. nidulans. Subsequently, Assis et al. (2015) identified seven other A. nidulans phosphatases involved in the regulation of cell cycle, development, and metabolism in response to glucose and alternative carbon sources. N. crassa genome encodes catalytic subunits for 30 protein phosphatase genes (Ghosh et al. 2014). These authors have characterized phenotypically in detail this null phosphatase collection by demonstrating that 91% of the mutants had defects during growth or asexual development or sexual development, whereas 29% have phenotypes in all three traits. Additionally, chemical sensitivity phenotypes were observed for 17 phosphatase null mutants and nine potential candidates for regulators of the p38 mitogen-activated protein kinase (MAPK) were identified. They have also recognized a phosphatase as a regulator of N. crassa female sexual development and Δcsp-1 and Δcsp-2, as important for regulation of conidiation and the circadian clock, respectively (Ghosh et al. 2014).
Previously we have identified 32 genes encoding catalytic subunits of protein phosphatases in the A. fumigatus genome (Winkelströter et al. 2015). Here, we further investigate the functions of protein phosphatases in A. fumigatus by generating a null mutant collection for the phosphatase catalytic subunit encoding genes. We were able to construct 24 viable phosphatase null mutants and their growth defects were analyzed, showing that the phosphatase mutants had a great deal of functional redundancy. Several phosphatase mutants had altered sensitivity to cell wall–damaging agents and oxidative and unfolded protein response, stressing chemicals, in addition to geldanamycin (GEL), a heat shock protein 90 (Hsp90) inhibitor. Subsequently, a group of protein phosphatases that possibly played a role in the HOG (high osmolarity glycerol response) pathway was assessed by measuring phosphorylation of the p38 MAPK (SakA) and the expression of osmo-dependent genes. In addition, several phosphatases were shown to be involved in the regulation of virulence factors, including iron assimilation and gliotoxin production/resistance. The phosphatase null mutant collection and these results provide a resource to dissect the signaling pathways and mechanisms involved in regulating virulence and stress tolerance in A. fumigatus. This deeper understanding of how virulence mechanisms are coordinated will have both biotechnological and biomedical implications.
Materials and Methods
Strains, media, and growth conditions
The A. fumigatus parental strains used in this study were CEA17 (control strain) and CEA17-80 (ku80-; pyrG-; this strain was used as a recipient strain for the deletion of all phosphatase genes), and Af293 (the parental strain for ΔsakA, ΔmpkC, and ΔsakA ΔmpkC). Media were of two basic types: a complete medium with three variants, YAG (2% glucose, 0.5% yeast extract, 2% agar, trace elements), YUU (YAG supplemented with 1.2 g/liter each of uracil and uridine), and liquid YG or YUU medium of the same compositions (but without agar), and a modified minimal medium (MM: 1% glucose, original high nitrate salts, trace elements, 2% agar, pH 6.5) was also used. Trace elements, vitamins, and nitrate salts are described by Käfer (1977). Expression of genes under the control of niiA promoter was regulated by nitrate source: repression on a modified minimal medium (MMM: 1% w/v glucose, 2% w/v agar) plus ammonium tartrate (50 mM) and induction on AMM plus sodium nitrate (10 mM). Strains were grown at 37° unless indicated otherwise. For the experiments of iron starvation, we have grown the strains in MM for 24 hr at 37° and transferred the mycelia to AMM (glucose 1%, salt solution without FeSO40.7H2O, and sodium nitrate 70 mM) plus BPS 200 µM [Bathophenanthrolinedisulfonic acid (4,7-diphenyl-1,10-phenanthrolinedisulfonic acid)] and 3-(2-pyridyl)-5,6-bis(4-phenylsulfonic acid)-1,2,4-triazine (ferrozine) 300 µM for 1 or 2 hr at 37°. For the experiments of iron excess, the strains were grown in AMM medium plus BPS 200 µM and ferrozine 300 µM for 24 hr and transferred to AMM plus FeSO40.7H2O 200 µM for 1 or 2 hr at 37°. The A. fumigatus phosphatase mutants constructed in this study are presented in Table 1. The A. fumigatus MAP kinase mutants ΔsakA, ΔmpkC, ΔmpkC ΔsakA, and ΔmpkA were constructed by Hagiwara et al. (2013, 2014) and Valiante et al. (2008), respectively.
Aspergillus fumigatus phosphatase gene families
| Familya . | Subfamilyb . | Class/Domainc . | A. fumigatus ID Genes . | A. fumigatus Proteins . | Effect of the Deletion on A. fumigatus . | A. nidulans Genes . | S. cerevisiae Protein . |
|---|---|---|---|---|---|---|---|
| S/T | PPP | PP2Ac | Afu5g12010 | PphA | Viable | AN0103 | Pph3p |
| S/T | PPP | PP2Ac | Afu5g11370 | PpgA | NDd | AN0164 | Ppg1p |
| S/T | PPP | PP2Ac | Afu1g04950 | GlcA | Lethal | AN0410 bimG | Glc7p |
| S/T | PPP | PP2Ac | Afu6g11470 | SitA | Viable | AN0504 sitA | Sit4P |
| S/T | PPP | PP2Ac | Afu2g03950 | PpzA | Viable | AN3793 | Ppz1p |
| S/T | PPP | PP2Ac | Afu6g10830 | PphB | Lethal | AN6391 pphA | Pph21p |
| S/T | PPP | PP2Ac | Afu5g06700 | PptA | Viable | AN10281 | Ppt1p |
| S/T | PPP | PP2Bc | Afu5g09360 | CalA/CnaA | Viable | AN8820 cnaA | Cmp2p |
| S/T | PPM | PP2Cc | Afu1g15800 | PtcA | Viable | AN0914 | Ptc6p |
| S/T | PPM | PP2Cc | Afu1g09280 | PtcB | Viable | AN1358 | Ptc2p |
| S/T | PPM | PP2Cc | Afu8g04580 | PpmA | Viable | AN1467 | −/− |
| S/T | PPM | PP2Cc | Afu5g13740, | PtcD, | Both viable | AN2472 | Ptc2p |
| Afu2g03890 | PtcE | ||||||
| S/T | PPM | PP2Cc | Afu1g06860 | PtcF | Viable | AN5722 | Ptc5p |
| S/T | PPM | PP2Cc | Afu5g13340 | PtcG | Viable | AN6892 | Ptc1p |
| S/T | PPM | PP2Cc | Afu4g00720 | PtcH | Viable | AN2472 | Ptc1p |
| S/T | Asp-based | HAD | Afu1g09460 | NemA | Viable | AN1343 | Nem1p |
| S/T | Asp-based | HAD | Afu3g11410 | FcpA | Lethal | AN2902 | Fcp1p |
| S/T | Asp-based | HAD | Afu1g04790 | PsrA | Viable | AN10077 | Psr1p |
| PTP | Dual-specificity | DSPc | Afu5g11690 | PpsA | Viable | AN0129 | −/− |
| PTP | Dual-specificity | DSPc | Afu4g07080 | DspC | Lethal | AN4419 | −/− |
| PTP | Dual-specificity | DSPc | Afu2g02760 | DspD | Viable | AN4544 | −/− |
| PTP | Dual-specificity | DSPc | Afu3g12250 | CdcA | Viable | AN5057 | Cdc14p |
| PTP | Dual-specificity | DSPc | Afu1g13040 | DspA | Viable | AN10138 | −/− |
| PTP | Dual-specificity | DSPc | Afu1g03540 | DspB | Viable | AN4057 | −/− |
| PTP | Classical | PTPc | Afu3g10970 | PtpB | Viable | AN4896 | Ptp1p |
| PTP | Classical | PTPc | Afu4g04710 | PypA | Viable | AN6982 | Ptp2p |
| PTP | LMW-PTP | LMWPc | Afu2g01880 | LtpA | Viable | AN10570 | Ltp1p/Yvh1p |
| SSU72 | SSU72 | SSU72 | Afu2g03760 | SsuA | NDd | AN3810 | Ssu72p |
| PTP | CDC-25 type | CDC25 | Afu6g08200 | NimT | NDd | AN3941 nimT | −/− |
| PTP | — | Y-fosfatase | Afu4g07000 | YphA | Viable | AN4426 | −/− |
| PTP | — | Y-fosfatase 3 | Afu6g06650 | PtyA | Viable | AN5767 | Ptc7p |
| Familya . | Subfamilyb . | Class/Domainc . | A. fumigatus ID Genes . | A. fumigatus Proteins . | Effect of the Deletion on A. fumigatus . | A. nidulans Genes . | S. cerevisiae Protein . |
|---|---|---|---|---|---|---|---|
| S/T | PPP | PP2Ac | Afu5g12010 | PphA | Viable | AN0103 | Pph3p |
| S/T | PPP | PP2Ac | Afu5g11370 | PpgA | NDd | AN0164 | Ppg1p |
| S/T | PPP | PP2Ac | Afu1g04950 | GlcA | Lethal | AN0410 bimG | Glc7p |
| S/T | PPP | PP2Ac | Afu6g11470 | SitA | Viable | AN0504 sitA | Sit4P |
| S/T | PPP | PP2Ac | Afu2g03950 | PpzA | Viable | AN3793 | Ppz1p |
| S/T | PPP | PP2Ac | Afu6g10830 | PphB | Lethal | AN6391 pphA | Pph21p |
| S/T | PPP | PP2Ac | Afu5g06700 | PptA | Viable | AN10281 | Ppt1p |
| S/T | PPP | PP2Bc | Afu5g09360 | CalA/CnaA | Viable | AN8820 cnaA | Cmp2p |
| S/T | PPM | PP2Cc | Afu1g15800 | PtcA | Viable | AN0914 | Ptc6p |
| S/T | PPM | PP2Cc | Afu1g09280 | PtcB | Viable | AN1358 | Ptc2p |
| S/T | PPM | PP2Cc | Afu8g04580 | PpmA | Viable | AN1467 | −/− |
| S/T | PPM | PP2Cc | Afu5g13740, | PtcD, | Both viable | AN2472 | Ptc2p |
| Afu2g03890 | PtcE | ||||||
| S/T | PPM | PP2Cc | Afu1g06860 | PtcF | Viable | AN5722 | Ptc5p |
| S/T | PPM | PP2Cc | Afu5g13340 | PtcG | Viable | AN6892 | Ptc1p |
| S/T | PPM | PP2Cc | Afu4g00720 | PtcH | Viable | AN2472 | Ptc1p |
| S/T | Asp-based | HAD | Afu1g09460 | NemA | Viable | AN1343 | Nem1p |
| S/T | Asp-based | HAD | Afu3g11410 | FcpA | Lethal | AN2902 | Fcp1p |
| S/T | Asp-based | HAD | Afu1g04790 | PsrA | Viable | AN10077 | Psr1p |
| PTP | Dual-specificity | DSPc | Afu5g11690 | PpsA | Viable | AN0129 | −/− |
| PTP | Dual-specificity | DSPc | Afu4g07080 | DspC | Lethal | AN4419 | −/− |
| PTP | Dual-specificity | DSPc | Afu2g02760 | DspD | Viable | AN4544 | −/− |
| PTP | Dual-specificity | DSPc | Afu3g12250 | CdcA | Viable | AN5057 | Cdc14p |
| PTP | Dual-specificity | DSPc | Afu1g13040 | DspA | Viable | AN10138 | −/− |
| PTP | Dual-specificity | DSPc | Afu1g03540 | DspB | Viable | AN4057 | −/− |
| PTP | Classical | PTPc | Afu3g10970 | PtpB | Viable | AN4896 | Ptp1p |
| PTP | Classical | PTPc | Afu4g04710 | PypA | Viable | AN6982 | Ptp2p |
| PTP | LMW-PTP | LMWPc | Afu2g01880 | LtpA | Viable | AN10570 | Ltp1p/Yvh1p |
| SSU72 | SSU72 | SSU72 | Afu2g03760 | SsuA | NDd | AN3810 | Ssu72p |
| PTP | CDC-25 type | CDC25 | Afu6g08200 | NimT | NDd | AN3941 nimT | −/− |
| PTP | — | Y-fosfatase | Afu4g07000 | YphA | Viable | AN4426 | −/− |
| PTP | — | Y-fosfatase 3 | Afu6g06650 | PtyA | Viable | AN5767 | Ptc7p |
Family abbreviations: S/T, serine/threonine; PTP, protein tyrosine phosphatase.
Subfamily abbreviations: PPP, phosphoprotein phosphatase; PPM, Mg2+ or Mn2+-dependent protein phosphatase; Asp-based, aspartate-based phosphatase; LMW-PTP, low-molecular-weight protein tyrosine phosphatase; CDC25 type, cell division cycle 25 type; SSU72, C-terminal domain RNA Pol II phosphatase.
Class/domain abbreviations: PP2Ac, protein phosphatase 2 A catalytic subunit; PP2Bc, protein phosphatase 2B catalytic subunit; PP2Cc, protein phosphatase 2C catalytic subunit; HAD, haloacid dehalogenase; PTPc, protein tyrosine phosphatase catalytic subunit; DSPc, dual-specificity phosphatase catalytic subunit; LMWPc, low-molecular-weight phosphatase catalytic subunit; CDC25, cell division cycle; SSU72, C-terminal domain RNA polymerase II phosphatase; Y-phosphatase 3, tyrosine phosphatase 3.
Not determined.
| Familya . | Subfamilyb . | Class/Domainc . | A. fumigatus ID Genes . | A. fumigatus Proteins . | Effect of the Deletion on A. fumigatus . | A. nidulans Genes . | S. cerevisiae Protein . |
|---|---|---|---|---|---|---|---|
| S/T | PPP | PP2Ac | Afu5g12010 | PphA | Viable | AN0103 | Pph3p |
| S/T | PPP | PP2Ac | Afu5g11370 | PpgA | NDd | AN0164 | Ppg1p |
| S/T | PPP | PP2Ac | Afu1g04950 | GlcA | Lethal | AN0410 bimG | Glc7p |
| S/T | PPP | PP2Ac | Afu6g11470 | SitA | Viable | AN0504 sitA | Sit4P |
| S/T | PPP | PP2Ac | Afu2g03950 | PpzA | Viable | AN3793 | Ppz1p |
| S/T | PPP | PP2Ac | Afu6g10830 | PphB | Lethal | AN6391 pphA | Pph21p |
| S/T | PPP | PP2Ac | Afu5g06700 | PptA | Viable | AN10281 | Ppt1p |
| S/T | PPP | PP2Bc | Afu5g09360 | CalA/CnaA | Viable | AN8820 cnaA | Cmp2p |
| S/T | PPM | PP2Cc | Afu1g15800 | PtcA | Viable | AN0914 | Ptc6p |
| S/T | PPM | PP2Cc | Afu1g09280 | PtcB | Viable | AN1358 | Ptc2p |
| S/T | PPM | PP2Cc | Afu8g04580 | PpmA | Viable | AN1467 | −/− |
| S/T | PPM | PP2Cc | Afu5g13740, | PtcD, | Both viable | AN2472 | Ptc2p |
| Afu2g03890 | PtcE | ||||||
| S/T | PPM | PP2Cc | Afu1g06860 | PtcF | Viable | AN5722 | Ptc5p |
| S/T | PPM | PP2Cc | Afu5g13340 | PtcG | Viable | AN6892 | Ptc1p |
| S/T | PPM | PP2Cc | Afu4g00720 | PtcH | Viable | AN2472 | Ptc1p |
| S/T | Asp-based | HAD | Afu1g09460 | NemA | Viable | AN1343 | Nem1p |
| S/T | Asp-based | HAD | Afu3g11410 | FcpA | Lethal | AN2902 | Fcp1p |
| S/T | Asp-based | HAD | Afu1g04790 | PsrA | Viable | AN10077 | Psr1p |
| PTP | Dual-specificity | DSPc | Afu5g11690 | PpsA | Viable | AN0129 | −/− |
| PTP | Dual-specificity | DSPc | Afu4g07080 | DspC | Lethal | AN4419 | −/− |
| PTP | Dual-specificity | DSPc | Afu2g02760 | DspD | Viable | AN4544 | −/− |
| PTP | Dual-specificity | DSPc | Afu3g12250 | CdcA | Viable | AN5057 | Cdc14p |
| PTP | Dual-specificity | DSPc | Afu1g13040 | DspA | Viable | AN10138 | −/− |
| PTP | Dual-specificity | DSPc | Afu1g03540 | DspB | Viable | AN4057 | −/− |
| PTP | Classical | PTPc | Afu3g10970 | PtpB | Viable | AN4896 | Ptp1p |
| PTP | Classical | PTPc | Afu4g04710 | PypA | Viable | AN6982 | Ptp2p |
| PTP | LMW-PTP | LMWPc | Afu2g01880 | LtpA | Viable | AN10570 | Ltp1p/Yvh1p |
| SSU72 | SSU72 | SSU72 | Afu2g03760 | SsuA | NDd | AN3810 | Ssu72p |
| PTP | CDC-25 type | CDC25 | Afu6g08200 | NimT | NDd | AN3941 nimT | −/− |
| PTP | — | Y-fosfatase | Afu4g07000 | YphA | Viable | AN4426 | −/− |
| PTP | — | Y-fosfatase 3 | Afu6g06650 | PtyA | Viable | AN5767 | Ptc7p |
| Familya . | Subfamilyb . | Class/Domainc . | A. fumigatus ID Genes . | A. fumigatus Proteins . | Effect of the Deletion on A. fumigatus . | A. nidulans Genes . | S. cerevisiae Protein . |
|---|---|---|---|---|---|---|---|
| S/T | PPP | PP2Ac | Afu5g12010 | PphA | Viable | AN0103 | Pph3p |
| S/T | PPP | PP2Ac | Afu5g11370 | PpgA | NDd | AN0164 | Ppg1p |
| S/T | PPP | PP2Ac | Afu1g04950 | GlcA | Lethal | AN0410 bimG | Glc7p |
| S/T | PPP | PP2Ac | Afu6g11470 | SitA | Viable | AN0504 sitA | Sit4P |
| S/T | PPP | PP2Ac | Afu2g03950 | PpzA | Viable | AN3793 | Ppz1p |
| S/T | PPP | PP2Ac | Afu6g10830 | PphB | Lethal | AN6391 pphA | Pph21p |
| S/T | PPP | PP2Ac | Afu5g06700 | PptA | Viable | AN10281 | Ppt1p |
| S/T | PPP | PP2Bc | Afu5g09360 | CalA/CnaA | Viable | AN8820 cnaA | Cmp2p |
| S/T | PPM | PP2Cc | Afu1g15800 | PtcA | Viable | AN0914 | Ptc6p |
| S/T | PPM | PP2Cc | Afu1g09280 | PtcB | Viable | AN1358 | Ptc2p |
| S/T | PPM | PP2Cc | Afu8g04580 | PpmA | Viable | AN1467 | −/− |
| S/T | PPM | PP2Cc | Afu5g13740, | PtcD, | Both viable | AN2472 | Ptc2p |
| Afu2g03890 | PtcE | ||||||
| S/T | PPM | PP2Cc | Afu1g06860 | PtcF | Viable | AN5722 | Ptc5p |
| S/T | PPM | PP2Cc | Afu5g13340 | PtcG | Viable | AN6892 | Ptc1p |
| S/T | PPM | PP2Cc | Afu4g00720 | PtcH | Viable | AN2472 | Ptc1p |
| S/T | Asp-based | HAD | Afu1g09460 | NemA | Viable | AN1343 | Nem1p |
| S/T | Asp-based | HAD | Afu3g11410 | FcpA | Lethal | AN2902 | Fcp1p |
| S/T | Asp-based | HAD | Afu1g04790 | PsrA | Viable | AN10077 | Psr1p |
| PTP | Dual-specificity | DSPc | Afu5g11690 | PpsA | Viable | AN0129 | −/− |
| PTP | Dual-specificity | DSPc | Afu4g07080 | DspC | Lethal | AN4419 | −/− |
| PTP | Dual-specificity | DSPc | Afu2g02760 | DspD | Viable | AN4544 | −/− |
| PTP | Dual-specificity | DSPc | Afu3g12250 | CdcA | Viable | AN5057 | Cdc14p |
| PTP | Dual-specificity | DSPc | Afu1g13040 | DspA | Viable | AN10138 | −/− |
| PTP | Dual-specificity | DSPc | Afu1g03540 | DspB | Viable | AN4057 | −/− |
| PTP | Classical | PTPc | Afu3g10970 | PtpB | Viable | AN4896 | Ptp1p |
| PTP | Classical | PTPc | Afu4g04710 | PypA | Viable | AN6982 | Ptp2p |
| PTP | LMW-PTP | LMWPc | Afu2g01880 | LtpA | Viable | AN10570 | Ltp1p/Yvh1p |
| SSU72 | SSU72 | SSU72 | Afu2g03760 | SsuA | NDd | AN3810 | Ssu72p |
| PTP | CDC-25 type | CDC25 | Afu6g08200 | NimT | NDd | AN3941 nimT | −/− |
| PTP | — | Y-fosfatase | Afu4g07000 | YphA | Viable | AN4426 | −/− |
| PTP | — | Y-fosfatase 3 | Afu6g06650 | PtyA | Viable | AN5767 | Ptc7p |
Family abbreviations: S/T, serine/threonine; PTP, protein tyrosine phosphatase.
Subfamily abbreviations: PPP, phosphoprotein phosphatase; PPM, Mg2+ or Mn2+-dependent protein phosphatase; Asp-based, aspartate-based phosphatase; LMW-PTP, low-molecular-weight protein tyrosine phosphatase; CDC25 type, cell division cycle 25 type; SSU72, C-terminal domain RNA Pol II phosphatase.
Class/domain abbreviations: PP2Ac, protein phosphatase 2 A catalytic subunit; PP2Bc, protein phosphatase 2B catalytic subunit; PP2Cc, protein phosphatase 2C catalytic subunit; HAD, haloacid dehalogenase; PTPc, protein tyrosine phosphatase catalytic subunit; DSPc, dual-specificity phosphatase catalytic subunit; LMWPc, low-molecular-weight phosphatase catalytic subunit; CDC25, cell division cycle; SSU72, C-terminal domain RNA polymerase II phosphatase; Y-phosphatase 3, tyrosine phosphatase 3.
Not determined.
Analysis of siderophore production by reverse-phase high-performance liquid chromatography
For siderophore production analysis, all strains were grown at 37° in AMM according to the method of Pontecorvo et al. (1953), with 2% (w/v) glucose and 20 mM glutamine as carbon and nitrogen sources, respectively. Trace elements were not supplemented with iron and all glassware was washed with concentrated HCl to remove free iron. Liquid cultures (50 ml) were grown in 250 ml conical flasks, inoculated with 108 conidia, at 200 rpm and 37° for 24–72 hr. Culture supernatants (triplicate) were ferrated by the addition of FeSO4 to a final concentration of 1.5 mM. Intracellular ferricrocin (FC) analysis was adapted from the work of Szigeti et al. (2014). Briefly, mycelia from 24- to 72-hr cultures were harvested and lyophilized and 50 mg mycelia from each strain (duplicate) was added to 750 µl deionized H2O in Eppendorf tubes and homogenized by bead beating (10 min) using tungsten beads. Lysates were centrifuged (10,000g, 10 min) and supernatants (200 µl) were removed and ferrated by addition of FeSO4 to a final concentration of 1.5 mM. All ferrated siderophores were analyzed by reverse-phase high-performance liquid chromatography (RP-HPLC) with DAD (Agilent 1200 system) using a C18 RP-HPLC column (Agilent Zorbax Eclipse XDB-C18 Semi-Prep; 5 µm, 9.4 × 250 mm) at a flow rate of 2 ml/min. Ferrated fusarinine C (FusC), triacetylfusarinine C (TAFC), and FC were detected at 440 nm. Purified standards of FusC and TAFC were used to determine the respective retention times. The milli absorbance unit (mAU) areas of all siderophores were determined for each sample.
Analysis of gliotoxin production by LC-MS/MS
For gliotoxin production analysis, all strains were grown at 37° in Czapek-Dox minimal medium. Liquid cultures (50 ml) were conducted in 250-ml conical flasks, inoculated with 108 conidia, at 200 rpm and 37° for 72 hr. Culture supernatants were analyzed by LC-MS/MS on an Agilent 6340 Ion Trap mass spectrometer (Dolan et al. 2014). Briefly, samples were organically extracted using chloroform (1:1) and the organic layer was evaporated under vacuum and resuspended in methanol. Samples were diluted 1/10 in 0.1% (v/v) formic acid and spin-filtered (Costar Spin-X) prior to LC-MS analysis; 1 µl of each sample was injected onto a Zorbax 300 SB C18 Nano-HPLC Chip (150 mm × 75 μm) with 0.1% (v/v) formic acid at a flow rate of 4 μl/min. Metabolites were eluted using an acetonitrile gradient with a post-run time of 5 min. A commercial standard of gliotoxin (Sigma-Aldrich) was utilized to confirm the retention time and fragmentation pattern of gliotoxin in culture supernatants.
Phylogenetic analysis
The protein sequences were obtained from the A. fumigatus genome database (http://www.aspgd.org) and the S. cerevisiae genome database (http://www.yeastgenome.org) (Supporting Information, File S1). The phylogenetic analysis was performed by using MEGA version 6 (Tamura et al. 2013). The alignment was performed with CulstalW and manually curated. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches (Felsenstein 1985). The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site (Zuckerkandl and Pauling 1965). The analysis involved 61 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1353 positions in the final dataset.
DNA manipulations and construction of the A. fumigatus mutants
The cassettes for gene replacement were constructed by in vivo recombination in S. cerevisiae as previously described by Colot et al. (2006). Approximately 1.5 kb from the 5′-untranslated region (UTR) and 3′-UTR flanking region of the targeted genes were selected for primer design. The primers 5F and 3R contained a short sequence homologous to the multiple cloning site (MCS) of the pRS426 plasmid. Both the 5- and 3-UTR fragments were PCR-amplified from A. fumigatus genomic DNA (gDNA). The pyrG inserted into the gene replacement cassettes was amplified from pCDA21 plasmid and was used to generate a marker for prototrophy in the mutant strains. Each fragment along with the BamHI/EcoRI cut pRS426 plasmid were transformed into the S. cerevisiae strain SC94721 using the lithium acetate method (Schiestl and Gietz 1989). The transformant DNA was extracted according to Goldman et al. (2003). The cassette was PCR-amplified from the plasmids utilizing TaKaRa Ex Taq DNA Polymerase (Clontech Takara Bio) and used for A. fumigatus transformation. Southern blot was performed as described by Sambrook et al. (1989) aiming to demonstrate that the transformation cassettes had integrated homologously at the targeted A. fumigatus loci. DNA fragments were labeled with 32P-α-dCTP using the RTS Rad Prime DNA labeling System kit (Invitrogen).
The promoter replacement strategy was utilized when the entire gene deletion was not possible. The DNA cassette containing niiA was constructed by transformation of the S. cerevisiae strain SC9721 with the PCR-amplified fragments of an approximately 1.5-kb 5′ flank, the ORF, and the pyrG::niiA fragment. The ORF is under the control of the A. fumigatus niiA promoter after homologous integration of the translation produces an N-terminal fusion protein. All the transformants were confirmed by PCR using specific primers and by checking if the promoter from niiA (encoding a nitrite reductase) is induced by sodium nitrate and repressed by ammonium tartrate (Punt et al. 1991). The primers and probes used above are described in Table S1 and Table S2. All the Southern blots, PCRs, and the corresponding strategies to evaluate if the phosphatase genes were either deleted or replaced by niiA are shown in Figure S1.
Phenotypic assays
The phenotypes of the deletion mutants were evaluated either by radial growth or by assessing the initial growth of a droplet of conidia from a serial dilution, at different temperatures, in the presence or absence of oxidative and osmotic stressing agents plus reagents that cause cell wall or DNA damage. Drop out experiments were performed using 5 µl of a 10-fold dilution series starting at a concentration of 2×107 for the wild-type and mutant strains spotted on different growth media and grown for 48 hr at 37°. Additionally, we have performed dry weight experiments by growing different strains for 48 hr at 37° and washing and lyophilizing the mycelia.
Immunoblot analysis
Detection of SakA phosphorylation by Western blotting was performed as described by Hagiwara et al. (2013) with slight modifications. Briefly, A. fumigatus conidia were inoculated into liquid YPD (1% yeast extract, 1% polypeptone, and 1% glucose) and cultured for 16 hr prior to addition or not (control) of 1/2 volume 3 M sorbitol (final concentration: 1 M). Mycelia were harvested, frozen in liquid nitrogen, and smashed with 0.5-mm glass beads in protein extraction buffer containing protease inhibitors. The suspension was centrifuged and the supernatant was boiled with an appropriate sample buffer. The protein concentration was determined using a Pierce BCA Protein Assay Kit-Reducing Agent Compatible (Pierce, Rockford, IL).
The same amount (10 µg) of protein was loaded onto NuPAGE Novex Bis-Tris 4–12% gel (Invitrogen). Proteins were separated with NuPAGE system (Invitrogen) and blotted using iBlot gel transfer system (Invitrogen). To detect SakA and phosphorylated SakA proteins, a rabbit polyclonal IgG antibody against Hog1 y-215 (Santa Cruz Biotechnology, Santa Cruz, CA) and a rabbit polyclonal IgG antibody against dually phosphorylated p38 MAPK (Cell Signaling Technology, Beverly, MA) were used, respectively. To detect these signals on blotted membranes, the ECL Prime Western Blotting Detection System (GE Healthcare, Little Chalfont, UK) and LAS1000 (FUJIFILM, Tokyo, Japan) were used.
RNA and cDNA preparation
Mycelia were harvested and frozen in liquid nitrogen, and total RNA was isolated using the FastRNA Pro Red Kit (MP Biomedicals, Santa Ana, CA). To obtain cDNA pools from the total RNA, the possible contaminating genomic DNA was removed and reverse-transcription was performed using the ReverTra Ace qPCR RT Master Mix with gDNA remover (Toyobo, Osaka, Japan).
Quantitative real-time RT-PCR
Real-time RT-PCR analysis was performed using the 7300 system (Life Technologies Corporation, Carlsbad, CA) with SYBR Green detection as described previously (Hagiwara et al. 2013). Briefly, the Thunderbird SYBR qPCR Mix was used for reaction mixture preparation. The primer sets for the analyses are listed in Table S1. The relative expression ratios were calculated by the ΔCt method. The actin gene was used as a normalization reference for target gene expression level, and wild-type before sorbitol treatment was set as the calibrator in each experiment. Each sample was tested in triplicate.
Results
Phenotypic characterization of A. fumigatus phosphatase null mutants
Previously, by using a combination of bioinformatics approaches, we were able to identify all the putative protein phosphatases in the A. fumigatus genome (Winkelströter et al. 2015). This analysis identified 32 A. fumigatus phosphatase catalytic subunit encoding genes in accordance with the A. fumigatus genome database (www.aspgd.org), and they were named following their S. cerevisiae homologues (Table 1 and Figure S2). These phosphatases were classified as 19 S/T members (8 PPP, 8 PPM, and 3 Asp-based subfamily members), 11 PTP members (6 dual-specificity, 2 classical, 1 LMW-PTP, 1 Cdc25-type, and 1 SSU72), and 2 fungal-specific phosphatases in the PTP family (YphA and PtyA) (Table 1) (Winkelströter et al. 2015).
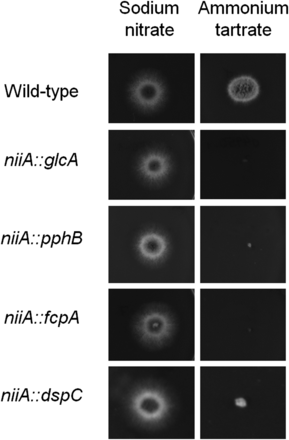
To gain a deeper insight into the function of the identified phosphatases, we attempted to construct null mutants for the 31 phosphatase encoding genes (because A. fumigatus calA gene encoding the catalytic subunit of calcineurin has already been deleted) (Steinbach et al. 2007; Da Silva Ferreira et al. 2007). We were able to construct 24 null mutants lacking a single phosphatase (Table 1). The inability to generate null mutants for the remaining seven genes could have been due to the fact that these were essential genes. Thus, conditional mutants were constructed for these genes by replacing the endogenous promoters with the niiA promoter (from the A. fumigatus nitrite reductase gene). The niiA promoter is induced by sodium nitrate and repressed by ammonium tartrate. Four of these genes (glcA, pphB, fcpA, and dspA) were shown to be essential (Figure 1). The functionality of the remaining three strains could not be assessed as the construction of null or conditional mutants (by using either niiA or alcA from the alcohol dehydrogenase gene and promoters) were unsuccessful for ppgA (Posas et al. 1993), ssuA (Sun and Hampsey 1996), and nimT (Russell et al. 1989). In A. nidulans, the homologous ppgA and ssuA null mutants have reduced fitness (Son and Osmani 2009), whereas the nimT homolog, also nimT, is an essential A. nidulans gene involved in mitosis progression (Son and Osmani 2009). Therefore, these genes are likely to also be essential genes in A. fumigatus.
Essential A. fumigatus phosphatase encoding genes. The wild-type, niiA::glcA, niiA::pphB, niiA::fcpA, and niiA::dspCA were grown for 48 hr at 37° on MM+sodium nitrate (induced) and MM+ammonium tartrate (repressed).
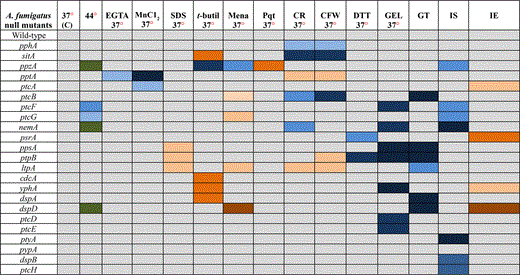
Growth of the 24 null mutants was compared to the wild-type strain in the following conditions: (1) different temperatures (30°, 37°, and 44°); (2) in media of different nutritional states [complete media (YAG), minimal media (MM), and fetal bovine serum (FBS)]; (3) during calcium starvation [ethylene glycol tetraacetic acid (EGTA)]; (4) for sensitivity to manganese chloride (MnCl2); (5) sodium dodecyl sulfate (SDS); (6) oxidative stress (t-butyl peroxide, menadione, and paraquat); (7) osmotic stress (NaCl and sorbitol); (8) cell wall–damaging agents [Congo Red (CR) and Calcofluor White (CFW)]; (9) unfolded protein response (UPR) [dithiotreitol (DTT)]; (10) GEL inhibition; (11) iron assimilation; and (12) gliotoxin production/sensitivity (Table 2, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, Figure 9, Figure 10). No dramatic differences in growth for the phosphatase null mutants were observed in comparison to the wild-type strain (Table 2 and Figure S3), except for the ΔptcF and ΔptcG mutants that had reduced growth at 44° (Table 2 and Figure S3). In addition, the ΔppzA, ΔnemA, and ΔdspD mutants had reduced conidiation at 44° (Table 2 and Figure S3).
Growth phenotypes of phosphatase null mutants compared to the wild-type strain
 |
 |
Abbreviations/growth: C, control/solid or liquid medium; EGTA, ethylene glycol tetraacetic acid/solid medium; SDS, sodium dodecyl sulfate/solid medium; t-butyl, t-butyl peroxide/liquid medium; Mena, menadione/liquid medium; Pqt, paraquat/liquid medium; CR, Congo Red/solid medium; CFW, Calcofluor White/solid medium; DTT, dithiotreitol/liquid medium; GEL, geldanamycin/liquid medium; GT, Gliotoxin/solid medium; IS, iron starvation/liquid medium; and IE, iron excess/liquid medium. Colors are explained in the following chart.
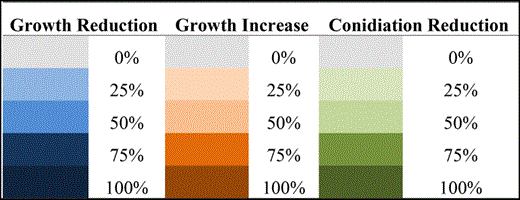
 |
 |
Abbreviations/growth: C, control/solid or liquid medium; EGTA, ethylene glycol tetraacetic acid/solid medium; SDS, sodium dodecyl sulfate/solid medium; t-butyl, t-butyl peroxide/liquid medium; Mena, menadione/liquid medium; Pqt, paraquat/liquid medium; CR, Congo Red/solid medium; CFW, Calcofluor White/solid medium; DTT, dithiotreitol/liquid medium; GEL, geldanamycin/liquid medium; GT, Gliotoxin/solid medium; IS, iron starvation/liquid medium; and IE, iron excess/liquid medium. Colors are explained in the following chart.

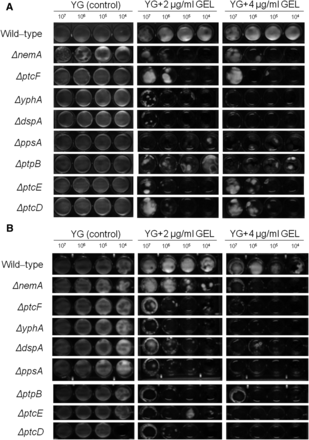
The A. fumigatus phosphatase mutants that were sensitive to GEL (Geldanamycin). Ten-fold conidial dilutions (107 to 104) of the wild-type and phosphatase null mutants were grown in MM in the absence or presence of different GEL concentrations for 48 hr at 37° (A) and 30° (B).
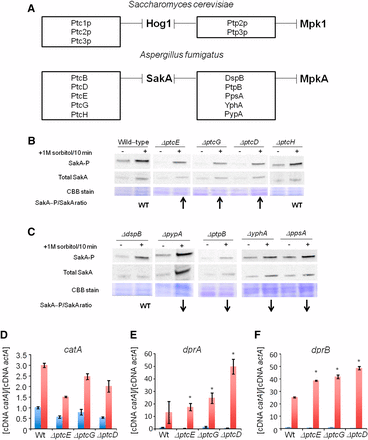
Identification of putative SakA phosphatase null mutants with increased SakA phosphorylation. (A) The putative A. fumigatus SakA and dual SakA/MpkA phosphatase homologs. Upper panel shows the S. cerevisiae Hog1p and dual Hog1p/Mpk1 phosphatases and the lower panel shows the putative A. fumigatus SakA and dual SakA/MpkA phosphatases. These data were based on the phylogenetic analysis shown in Figure S2. (B) and (C) Immunoblot analysis for SakA phosphorylation in response to osmotic stress. The wild-type and the phosphatase null mutants were grown for 18 hr at 37°. Then, sorbitol (1 M final concentration) was not added (control) or added for 10 min. The mycelium was harvested at the indicated times, and total proteins were extracted. Anti-phospho-p38 was used to detect the phosphorylation of SakA, and anti-Hog1p was used to detect the total SakA protein. A Coomassie Brilliant Blue (CBB)-stained gel is shown as a loading control for both gels. Signal intensities were quantified using the Image J software by dividing the intensity of SakA-P/SakA ratio. The experiment was repeated at least three times and a representative blot is shown. The “WT” signifies that the levels of SakA-P/total SakA on osmostress were similar to wild-type, whereas the arrows ↑ and ↓ that correspond to the levels of SakA-P/total SakA on osmostress were higher or lower than the wild-type, respectively. Phosphatase null mutants show higher expression of osmostress-dependent genes. The wild-type and the phosphatase null mutants were grown for 18 hr at 37°. Then, sorbitol (1 M final concentration) was added for 0 (control) and 10 min. The mycelium was harvested at the indicated times, and total RNA was extracted. The relative expression ratios of catA (D), dprA (E), and dprB (F) and actA (Afu6g04740, encoding the actin) were calculated by the ΔCt method. The results are the means (± SD) of three biological replicates (*P < 0.001, comparison of the treatments with the time zero control).
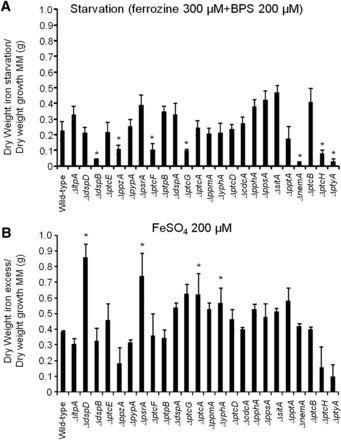
Identification of phosphatase null mutants involved in iron assimilation. The wild-type and the phosphatase null mutants were grown for 48 hr at 37° in minimal medium (MM), in AMM+ferrozine 300 μM+BPS 200 μM (iron starvation), or AMM+200 mM FeSO4 (iron excess). After this period, the mycelia were washed with sterile water and dried (*P < 0.001, comparison of the null mutants with the wild-type strain). The results are expressed as (A) dry weight of the strains grown on iron starvation divided by dry weight of the strains grown in MM and (B) dry weight of the strains grown on iron excess divided by dry weight of the strains grown in MM.
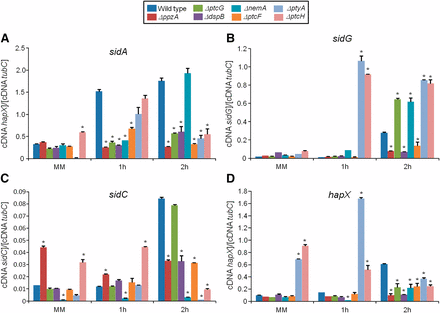
Phosphatase null mutants show altered expression of iron-dependent genes during iron starvation. The wild-type and the phosphatase null mutants were grown for 18 hr at 37° in MM. Then, the mycelia were transferred to AMM+ferrozine 300 μM+BPS 200 μM for 1 hr or 2 hr. The mycelium was harvested at the indicated times, and total RNA was extracted. The absolute quantitation of sidA (A), sidG (B), sidC (C), and hapX (D), and the normalizer tubC, was determined by a standard curve (i.e., CT values plotted against a logarithm of the DNA copy number). The results are the means (± SD) of three biological replicates (*P < 0.001, comparison of the null mutants with the wild-type strain).
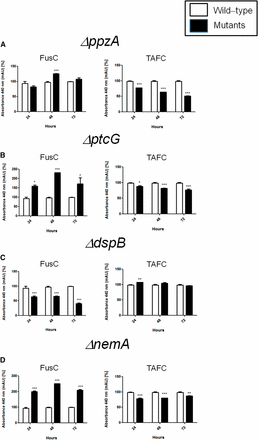
FusC and TAFC production in the wild-type and phosphatase null mutants that have reduced growth during iron starvation. The bars represent the integration areas of the FusC and TAFC peaks identified from culture supernatants analyzed by RP-HPLC (reverse-phase high-performance liquid chromatography). *P < 0.05; **P < 0.01; and ***P < 0.001.
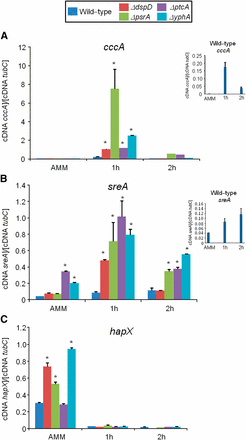
Phosphatase null mutants show altered expression of iron-dependent genes during iron excess. The wild-type and the phosphatase null mutants were grown for 18 hr at 37° in AMM+ferrozine 300 μM+BPS 200 μM. Then, the mycelia were transferred to AMM+200 mM FeSO4 for 1 hr or 2 hr. The mycelium was harvested at the indicated times, and total RNA was extracted. The absolute quantitation of cccA (A), sreA (B), hapX (C), and the normalizer tubC, was determined by a standard curve (i.e., CT values plotted against a logarithm of the DNA copy number). The insets in (A) and (B) show the results of the wild-type. The results are the means (± SD) of three biological replicates (*P < 0.001, comparison of the null mutants with the wild-type strain).
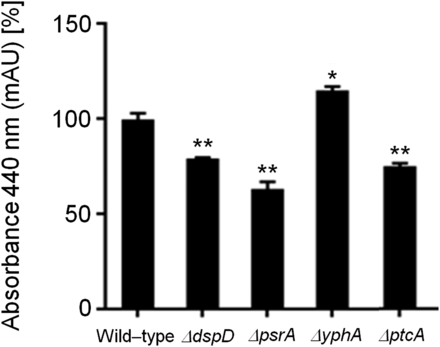
Ferricrocin (FC) production in the wild-type and phosphatase null mutants that have increased growth during iron excess. The bars represent the integration areas of the FC peaks identified from culture supernatants analyzed by RP-HPLC (reverse-phase high-performance liquid chromatography). The asterisks indicate statistical analysis using unpaired t-test (*P < 0.05 and **P < 0.01 when compared to the wild-type strain).
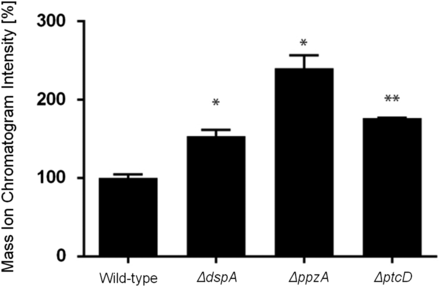
Gliotoxin (GT) detection in wild-type, ΔdspA, ΔppzA, and ΔptcD. The quantitative values represent the normalized intensity of GT determined by its diagnostic mass ion chromatograms (m/z 327). The bars represent the mean of three samples and error bars represent SE. The asterisks indicate statistical analysis using unpaired t-test (*P < 0.05 and **P < 0.01 when compared to the wild-type strain).
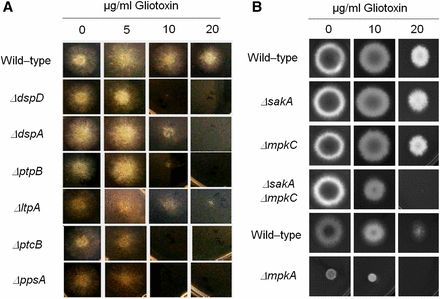
Identification of phosphatase null mutants more sensitive to gliotoxin. The wild-type and the phosphatase null mutants were grown for 72 hr at 37° in MM plus different concentrations of gliotoxin (A). Corresponding wild-type (strains Af293 for ΔsakA, ΔmpkC, and ΔsakA ΔmpkC, and CEA17 for ΔmpkA) and MAP kinase null mutants were grown for 72 hr at 37° in MM plus different concentrations of gliotoxin (A).
GEL inhibits Hsp90 by binding to its N-terminal ATP domain (Gorska et al. 2012). Eight of the phosphatase null mutants were more sensitive to GEL than the wild-type strain when grown at 37° (ΔnemA, ΔptcF, ΔyphA, ΔdspA, ΔppsA, ΔptpB, ΔptcE, and ΔptcD) (Figure 2A). The same results were observed at 45° (data not shown). To further evaluate if this GEL sensitivity was associated with growth temperature, GEL sensitivity was also assessed at 30° (Figure 2B). Seven null mutants remained sensitive to GEL, whereas the growth of the ΔnemA mutant was partially recovered at 30° (Figure 2B). This implied that these phosphatases were important for Hsp90 assembly and/or GEL was affecting other client proteins that interact with these phosphatases. Furthermore, it was possible that NemA participated in a signal transduction pathway associated with Hsp90 and thermo-tolerance. The GEL experiments could not be performed with the ΔptcB mutant due to adhesion problems and the fact that this mutant grew poorly in the microtiter plates (Winkelströter et al. 2015).
Taken together, these results strongly suggest that A. fumigatus phosphatases could have overlapping functions. However, several phosphatases were identified to have specific roles during oxidative and cell wall stressing conditions, whereas others influenced in Hsp90 function.
A. fumigatus HOG response phosphatases
Unexpectedly, none of phosphatase null mutants were very sensitive to osmotic stress either in liquid or in solid media, suggesting the existence of functional redundancy in the A. fumigatus osmotic stress pathway. In S. cerevisiae, five phosphatases (Ptc1p, Ptc2p, Ptc3p, Ptp2p, and Ptp3p) dephosphorylate Hog1p (Saito and Posas 2012; Brewster and Gustin 2014), negatively regulating the kinase activity of the osmotic stress and cell wall integrity (CWI) pathways (Figure 3A). In A. fumigatus, 10 putative homologues of the proteins PtcB, PtcD, PtcE, PtcG, and PtcH (for S. cerevisiaePtc1–Ptc3p) and DspB, PtpB, PpsA, YphA, and PypA (for S. cerevisiaePtp2–Ptp3p) were identified (Figure 3A and Figure S1). Recently, we have identified PtcB as the HOG phosphatase important for A. fumigatus virulence (Winkelströter et al. 2015). Here, we determine which of the other nine phosphatases were involved in the HOG pathway in A. fumigatus. Accordingly, the amount and phosphorylation state of Hog1p homolog, SakA, were determined in the presence and absence of osmotic stress. The phosphorylation level of the SakA protein was determined using the anti-phospho-p38 MAPK (Thr180/Tyr182) and anti-Hog1 (y-215) antibodies (Figure 3, A and B). In the wild-type strain, SakA phosphorylation levels increase approximately twice post-transfer to sorbitol 1 M for 10 min (Figure 3B). Ten minutes was chosen because this was previously shown to be the time point with the highest SakA phosphorylation (Hagiwara et al. 2013). The ΔdspB, ΔptcH, and ΔppsA mutants demonstrated levels of SakA induction comparable to the wild-type strain, whereas the ΔptpB, ΔpypA, and ΔyphA mutants did not show any induction (Figure 3, B and C). Thus, the PtpB, PypA, and YphA phosphatases may perform functions important for the SakA phosphorylation. Interestingly, ΔptcE, ΔptcG, and ΔptcD mutants had increased levels of SakA phosphorylation (Figure 3B). Taken together, these results suggest that at least three other phosphatases besides PtcB influence the HOG pathway in A. fumigatus.
The genetic markers used to evaluate the induction of the HOG pathway in A. fumigatus included catA (catalase, Afu6g12180), dprA (dehydrin, Afu4g00860), and dprB (dehydrin, Afu6g12180). The mRNA accumulation of these genes was determined in the wild-type, ΔptcE, ΔptcG, and ΔptcD strains post-exposure to sorbitol (Figure 3, D–F). Catalase and dehydrin-like proteins play a role in oxidative, osmotic, and pH stress responses, and their expression is dependent on the HOG pathway (Wong Sak Hoi et al. 2011). On osmotic stress in the wild-type strain, catA, dprA, and dprB demonstrated an approximately 3-, 10-, and 25-fold increase in mRNA accumulation (Figure 3, D–F). In all the tested phosphatase mutants, dprA and dprB demonstrated higher mRNA accumulation than the wild-type strain (Figure 3, D–F). In contrast, catA mRNA accumulation was comparable to the wild-type in all tested null strains (Figure 3, D–F). These results are in accordance with the observed higher SakA phosphorylation levels, suggesting that the absence of these phosphatases results in an increased activation of the HOG pathway.
PtcB is an A. fumigatus HOG phosphatase (Winkelströter et al. 2015). Thus, to investigate the impact of the ΔptcB mutation on the transcriptional accumulation of ptcE, ptcG, and ptcD, and the subsequent redundancy of the system, the transcriptional regulation of these three phosphatase genes was assessed in the ΔptcB mutant after exposure to sorbitol (Figure S4). In the ΔptcB mutant, ptcE, ptcG, and ptcD mRNA accumulation was higher than in the wild-type strain (Figure S4). Taken together, these results indicate that PtcB and PtcD, together with PtcB (Winkelströter et al. 2015), were the major A. fumigatus HOG phosphatases, whereas PtcE and PtcG were minor contributors that may perform a greater role in the absence of either PtcB or PtcD.
Identification of phosphatases involved in iron assimilation
A. fumigatus cannot directly use human iron sources such as heme, ferritin, or transferrin (Schrettl and Haas 2011; Moore 2013; Haas 2014). It utilizes both reductive iron assimilation (RIA) and siderophore (low-molecular-mass ferric iron chelators)-mediated iron uptake during murine infection (Schrettl and Haas 2011; Moore 2013; Haas 2014). Two master transcription factors regulate iron assimilation, HapX (during starvation) and SreA (during iron replete or excess) (Schrettl and Haas 2011; Moore 2013; Haas 2014). A. fumigatus makes four types of siderophores. For iron uptake, it secretes fusarinine C (FsC) and triacetylfusarinine C (TAFC) and accumulates ferricrocin (FC) for hyphal and hydroxyferricrocin (HFC) for conidial iron distribution and storage (Schrettl and Haas 2011; Moore 2013; Haas 2014). HapX controls positively the transcription of several genes involved in FsC and TAFC biosynthesis, such as sidA (L-ornithine N5-oxygenase; the first committed step in siderophore biosynthesis), sidC [nonribosomal peptide synthetase (NRPS) involved in ferricrocin siderophore biosynthesis], and sidG (fusarinine C acetyltransferase) but represses sreA, whereas SreA induces cccA (an iron transporter of the vacuolar membrane, involved in vacuolar iron storage) but represses hapX (Schrettl and Haas 2011; Moore 2013; Haas 2014).
To identify phosphatases that regulate events involved in iron starvation or excess, we initially grew the wild-type and the phosphatase null mutants in conditions of iron replete, excess (200 mM FeSO4), and starvation for 48 hr (Figure 4). All the null phosphatase mutants have biomass growth comparable to the wild-type in iron replete conditions (data not shown). Seven mutants grew significantly less than the wild-type in iron starvation conditions: ΔppzA, ΔptcG, ΔdspB, ΔnemA, ΔptcF, ΔptcH, and ΔptyA (Figure 4A). Four mutants grew better than the wild-type in iron excess conditions: ΔdspD, ΔpsrA, ΔptcA, and ΔyphA (Figure 4B). The wild-type and the mutants were grown for 24 hr in iron replete or iron starvation conditions and then transferred to either iron starving or iron excess conditions for 1 or 2 hr (Figure 5 and Figure 7). As expected, in the wild-type strain during iron starvation conditions, sidA, sidC, sidG, and hapX exhibited increased mRNA accumulation (Figure 5, A–D). In contrast, in the seven mutants that were unable to grow during iron starvation conditions, there is a very complex pattern of gene expression. The sidA mRNA accumulation is lower in the mutants than in the wild-type strain, except for ΔptyA that has a delayed, but comparable, gene expression to the wild-type (Figure 5A). The sidG expression is much higher in ΔptyA, ΔptcH, ΔptcG, and ΔnemA than in the wild-type, whereas in the ΔppzA, ΔdspB, ΔptcF, and ΔptcH it was lower than in the wild-type (Figure 5B). All the mutants have reduction of sidC mRNA, except the ΔptcG mutant, which has an expression pattern similar to the wild-type (Figure 5C). All the mutants have reduced hapX mRNA accumulation, except ΔptyA and ΔptcH, which have a much earlier and higher (ΔptyA) accumulation than the wild-type (Figure 5D). We have also evaluated FusC and TAFC production in four of these mutants (randomly selected) (Figure 6). There is an increased and decreased production of FsC and TAFC, respectively, in all four mutants (Figure 6, A–D).
In the wild-type strain during iron excess conditions, the cccA and sreA have increased mRNA accumulation, whereas hapX has decreased mRNA accumulation after 1 and 2 hr (Figure 7, A–C, and insets in Figure 7, A and B). In contrast, most of the four mutants that have increased biomass in iron excess had an increased mRNA accumulation of cccA and sreA, and much lower hapX mRNA accumulation than in the wild-type strain (Figure 7, A–C). Surprisingly, only one of these four mutants, ΔyphA, exhibited higher intracellular FC production than the wild-type, whereas the three other mutants have lower FC production than the wild-type (Figure 8).
Taken together, these results strongly suggest that these phosphatases are involved directly or indirectly in post-translational modifications that affect the response to iron assimilation, impacting not only the transcription of several genes involved in the siderophore biosynthesis but also the siderophore production.
Recognition of phosphatase null mutants with higher gliotoxin production and sensitivity
Gliotoxin is an epidithiodioxopiperazine (ETP)-type fungal toxin that performs an important role in A. fumigatus virulence by inducing apoptosis in macrophages and modulating the immune response (Scharf et al. 2012). All the phosphatase null mutants produce gliotoxin levels comparable to the wild-type strain, except for the null mutants for ΔdspA, ΔptcD, and ΔppzA, which were able to produce 1.5-, 1.8-, and 2.4-fold more gliotoxin than the wild-type strain (Figure 9, Figure S5, Figure S6). The ΔdspD, ΔdspA, ΔptpB, ΔltpA, ΔptcB, and ΔppsA mutants showed increased sensitivity to gliotoxin than the wild-type strain (Figure 10A). Considering that four of these phosphatases, PtcD, PtpB, PpsA (shown here), and PtcB (Winkelströter et al. 2015) are HOG phosphatases (see Figure 3), we decided to investigate the contribution of different A. fumigatus MAP kinases to gliotoxin sensitivity (Figure 10B). There are four MAPKs in A. fumigatus: (1) MpkA (regulation of CWI signaling and pyomelanin formation); (2) MpkB (mating, putative pheromone signaling); (3) MpkC (regulation of conidium germination); and (4) SakA (the Hog1 ortholog that is involved in osmotic stress, carbon and nitrogen starvation, and regulation of conidium germination) (Xue et al. 2004, Reyes et al. 2006; May 2008; Valiante et al. 2008, 2009). The ΔsakA, ΔmpkC, ΔsakA ΔmpkC, and ΔmpkA strains were tested for gliotoxin sensitivity (Figure 10B; note that the parental strain of ΔmpkA is Af293, whereas for the other mutants CEA17 is the parental strain). The double mutant ΔsakA ΔmpkC was much more sensitive to gliotoxin than the single null mutants and the wild-type strains (Figure 10B). Taken together, these results suggest that SakA and MpkC kinase and phosphatase pathways are redundant and important for gliotoxin resistance.
Discussion
Protein phosphatases have been portrayed as important in virulence and pathogenicity in several human and pathogenic fungi (Erental et al. 2007; Di Stasio et al. 2009; Rispail et al. 2009; Feng et al. 2010; Jiang et al. 2011; Ariño et al. 2011; Adám et al. 2012; Du et al., 2013; Yang et al. 2013a,b; Shin et al. 2013; Muszkieta et al. 2014: Lee et al. 2014; Yu et al. 2014). The presented study of a collection of A. fumigatus protein phosphatase null mutants demonstrates the value of this novel biological resource in dissecting the signaling pathways involved in infection. Subsequently, this investigation revealed the importance of multiple phosphatases in regulating virulence traits, particularly osmotic stress resistance, iron assimilation, and gliotoxin production/sensitivity. We have reconstituted ΔptpB (Winkelströter et al. 2015), ΔppzA, ΔptcG (data not shown), and ΔsitA (V. L. P. Bom, unpublished data) strains and confirmed that the corresponding phenotypes observed here are only due to the phosphatase mutations. Before our investigation, there were only two previous studies reporting A. fumigatus phosphatase PhzA (protein phosphatase Z, here named PpzA) being involved in oxidative stress resistance (Leiter et al. 2012; Muszkieta et al. 2014) and the HOG phosphatase PtcB (Winkelströter et al. 2015).
The A. fumigatus genome contains 32 putative protein phosphatase catalytic subunit-encoding genes. However, not all genes could be deleted during the construction of the phosphatase null mutant collection, with phosphatases including 19 serine/threonine and 13 tyrosine phosphatases. Interestingly, fungi do not possess proper tyrosine kinases that phosphorylate tyrosine residues (Borkovich et al. 2004; Kosti et al. 2010). The current hypothesis for the presence of tyrosine phosphatases in fungal genomes, including A. fumigatus, is that tyrosine phosphatases have evolved before tyrosine kinases because serine/threonine kinases can phosphorylate tyrosine residues to a lesser extent, creating a target for the tyrosine phosphatases (Moorhead et al. 2007, 2009). Eight A. fumigatus phosphatases, PpmA (Afu8g04580), PpsA (Afu5g11690), DspC (Afu4g07080), DspD (Afu2g02760), DspA (Afu1g13040), DspB (Afu1g03540), NimT (Afu6g08200), and YphA (Afu4g07000) showed very low or no identity to other proteins in S. cerevisiae, animals, or plants, whereas putative homologues were identified in A. nidulans and N. crassa (except for PtyA that has no N. crassa homologue). This could indicate these phosphatases are fungal-specific. Similarly, in N. crassa two PTPs, pty-5 and pty-6, also appear to be fungal-specific phosphatases, whereas disruption of these phosphatases causes defects in fungal-specific traits, such as conidiation and resistance to fludioxonil (Ghosh et al. 2014). For the fungal-specific phosphatase mutants in A. fumigatus, we observed sensitivity to GEL and SDS (ΔppsA), reduced conidiation at 44° and sensitivity to menadione (ΔdspD), sensitivity to t-butyl (ΔdspA), involvement in the cell cycle (ΔnimT), and sensitivity to GEL and t-butyl (ΔyphA) (Table 2). Additional studies are necessary to evaluate the importance of these putative fungal-specific tyrosine phosphatases in filamentous fungi.
Through extensive screening for in vitro phenotypes using combinations of numerous compounds and conditions, it was possible to assign specific phenotypes for all the phosphatase null mutants, except ΔpypA. Some of these mutants have complex phenotypes, such as sensitivity to cell-damaging agents and reduced growth during iron starvation. This may reflect the complex net of signal transduction pathways that influence these traits and, accordingly, how these phosphatases are engaged in the activation and repression of different protein interactions. Interestingly, none of the phosphatase mutants displayed sensitivity to high osmolarity, suggesting the existence of functional redundancy. It remains to be investigated if the null phosphatase mutants could affect other aspects of the A. fumigatus biology, such as sexual cycle or other stages of the life cycle.
The phosphatase null mutant collection was screened using a chemical genomic approach with GEL, an inhibitor of Hsp90 (Gorska et al. 2012). The fungal Hsp90 interactome has numerous client proteins such as receptors, protein kinases, and transcription factors (Leach et al. 2012). It has been shown that Cdc37p, an Hsp90 co-chaperone, controls the functionality of the Hog1 and Mpk1 cascades in S. cerevisiae (Hawle et al. 2007; Yang et al. 2007), suggesting that Cdc37p acts as a regulator of MAPK signaling. Diezmann et al. (2012) used an equivalent chemical genomic screening approach to identify the C. albicans Hsp90 interaction network under diverse stress conditions. This study revealed that the chaperone interactome was dependent on the environment and that most of the 226 genetic interactors were important for growth only under specific conditions, suggesting that they operate downstream of Hsp90, as was the case for the MAPK Hog1. Leach et al. (2012) have observed that in C. albicans, Hsp90 interacts with and downregulates the heat shock transcription factor Hsf1, modulating short-term thermal adaptation, whereas long thermal adaptation depends on cross-talk between the Hog1, Mck1, and Cek1 MAPK cascades. In C. albicans, temperature affects the resistance of C. albicans to cell wall stresses but not osmotic stress, whereas Hsp90 depletion affects cell wall biogenesis by impairing the activation of its client proteins Mkc1 and Hog1, as well as Cek1 (Leach et al. 2012). These results indicate that in C. albicans Hsp90 modulates the short-term Hsf1-mediated activation of the classic heat shock response and coordinates this response with the long-term thermal adaptation process via Mkc1-, Hog1-, and Cek1-mediated cell wall remodeling (Leach et al. 2012). In A. fumigatus neither temperature nor sub-inhibitory concentrations of GEL had a dramatic influence on growth (data not shown). However, eight A. fumigatus phosphatase null mutants were sensitive to GEL. Further investigation is required to determine which signal transduction cascades are affected by Hsp90 inhibition in A. fumigatus.
S. cerevisiaeHog1 kinase is inactivated by the S/T phosphatases Ptc1, Ptc2, and Ptc3, and by the PTP phosphatases Ptp2 and Ptp3 (Jacoby et al. 1997; Wurgler-Murphy et al. 1997; Warmka et al. 2001; Saito and Tatebayashi 2004; Martín et al. 2005; Saito and Posas 2012). In S. cerevisiaePtp2 and Ptp3 also inactivate Mpk1 of the CWI pathway (see Figure 4) (Mattison et al. 1999; González et al. 2006). In N. crassa there are nine potential candidates for regulators of the p38 MAPK; among them are Δpph-8 and Δpty-3 (Ghosh et al. 2014). A. fumigatus PtcB, PtcD, PtcE, and PtyA are possible homologues of N. crassa pph-8 and pty-3, respectively. The presented study identified at least three putative Hog1 phosphatases in A. fumigatus that seem to have different influences on SakA activation state. Recently, we have identified PtcB as an additional SakA phosphatase (Winkelströter et al. 2015). Interestingly, PtcB also had a remarkable effect on the cell surface, conidia and germling adhesion, biofilm formation, and MpkA phosphorylation, suggesting PtcB is also involved in the CWI pathway (ten Cate et al. 2009; Cuéllar-Cruz et al. 2012). However, we were not able to observe any cell wall damage defects in the putative SakA phosphatases PtcD, PtcE, and PtcG. There are only few reports of Botrytis cinerea and Cryptococcus neoformans Hog1 phosphatases showing their significance in virulence and pathogenicity (Yang et al. 2013a,b; Lee et al. 2014), and the importance of A. fumigatus PtcD, PtcE, and PtcG on the survival in the host remains to be demonstrated.
We have identified 11 phosphatase null mutants as involved in iron metabolism, by showing their reduced or increased growth during iron starvation or excess, respectively. Accordingly, the mRNA accumulation of several genes related to iron starvation or excess and the siderophore production were anomalous when compared with the parental strain, suggesting these phosphatases are affecting transcriptional programs related to iron metabolism or the activity of enzymes involved in siderophore production. Both SreA and HapX transcription factors appear to be regulated post-translationally by iron (Schrettl and Haas 2011; Moore 2013; Haas 2014). Thus, it is possible these alterations in the transcriptional programs are related to post-translational modifications of these transcriptional master regulators performed by these phosphatases. Siderophore biosynthesis is important both for iron starvation and excess (Schrettl and Haas 2011; Moore 2013; Haas 2014). Their biosynthesis is affected by the available concentrations of the amino acid precursors arginine and ornithine and the ergosterol intermediate mevalonate (Schrettl and Haas 2011; Moore 2013; Haas 2014). Accordingly, it is also possible that the defects observed for these phosphatase mutants are dependent on the dynamics of the amino acid pools or changes in the concentration of mevalonate. However, we were not able to observe any change in the biomass of these mutants when they were grown either in starvation or in excess iron conditions in the presence of eflornithine (an ornithine analog) or lovastatin (an inhibitor of Hgm1, hydromethylglutaryl−CoA reductase, the enzyme that converts hydromethylglutaryl−CoA to mevalonate; data not shown).
Depending on the host immune status, gliotoxin is an important virulence factor for A. fumigatus; however, gliotoxin can also inhibit fungal development, including A. fumigatus growth (Scharf et al. 2012; Chotirmall et al. 2014; Carberry et al. 2012). The mediation of self-protection of A. fumigatus against gliotoxin is performed mainly by GliT, a gliotoxin sulfhydryl oxidase required for gliotoxin biosynthesis (Scharf et al. 2012; Chotirmall et al. 2014; Carberry et al. 2012; Schrettl et al. 2010). GliT is able to keep gliotoxin in the sulfur-bridged form, avoiding the generation of reactive oxygen species and of protein conjugates (Scharf et al. 2012; Chotirmall et al. 2014; Carberry et al. 2012). All the phosphatase null mutants produce comparable gliotoxin levels than the wild-type, except for the null mutants for DspA, PtcD, and PpzA that were able to produce between 1.5- and 2.4-fold more gliotoxin than the wild-type strain. We have observed six null phosphatase mutants as more sensitive to gliotoxin than the wild-type. These six phosphatases could participate in post-translational modifications that affect GliT activation or general mechanisms of gliotoxin detoxification. Interestingly, some of these phosphatases are putative MAP kinase phosphatases, such as PtpB, PpsA, PtcB, and PtcD. Consequently, we evaluated the gliotoxin sensitivity of three A. fumigatus MAP kinase null mutants. Our results strongly suggest that SakA and MpkC collaborate and are influential in mediating gliotoxin resistance in A. fumigatus.
This study presents the creation of an important novel biological resource for the dissection of the signal transduction pathways involved in pathogenicity. The value of this resource was subsequently demonstrated through the identification of the phosphatases responsible for the regulation of the SakA-mediated osmotic stress, iron assimilation, and gliotoxin resistance pathways. The continued investigation of the phosphatase mutant collection will reveal new connections among MAPK cascades and other signaling pathways involved in virulence. This could facilitate the design of novel strategies aiming to control this important disease.
Acknowledgments
We thank Drs. Axel Brakhage and Vito Valiante for providing us the A. fumigatus ΔmpkA mutant, the two anonymous reviewers and the editor for their comments and suggestions, and Dr. Elodie Bovier for help with the phylogenetic analysis. We also thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for providing financial support. S.K.D. is a recipient of an Irish Research Council Embark PhD Fellowship. Fungal research in the laboratory of S.D. is funded by Science Foundation Ireland Awards (PI/11/1188 & 12/IP/1695). HPLC and LC-MS facilities were funded by a competitive award from the Irish Higher Education Authority. Research exchanges between the laboratories of G.H.G., G.W.J., and S.D. were funded in part by SFI/12/ISCA/2494. R.A. is supported by a PhD scholarship from the Government of the Kingdom of Saudi Arabia.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.016766/-/DC1
Communicating editor: M. S. Sachs