-
PDF
- Split View
-
Views
-
Cite
Cite
Erika Anderson, Claire Burns, Miriam E Zolan, Global Gene Expression in Coprinopsis cinerea Meiotic Mutants Reflects Checkpoint Arrest, G3 Genes|Genomes|Genetics, Volume 2, Issue 10, 1 October 2012, Pages 1213–1221, https://doi.org/10.1534/g3.112.003046
Close - Share Icon Share
Abstract
The basidiomycete Coprinopsis cinerea is well-suited to studies of meiosis because meiosis progresses synchronously in 10 million cells within each mushroom cap. Approximately 20% of C. cinerea genes exhibit changing expression during meiosis, but meiosis and mushroom development happen concurrently and therefore differentially expressed genes might not be directly involved in meiotic processes. By using microarrays, we examined global gene expression across a meiotic time course in two mutants in which meiosis arrests but mushrooms develop normally. Genes differentially expressed in the mutants compared with the wild type are likely to be involved in meiosis and sporulation as opposed to mushroom development. In rad50-1, which arrests in late prophase, RNA abundance for a group of early meiotic genes remains high, whereas the expression of a group of late meiotic genes is never induced. In contrast, in msh5-22 (which fails to undergo premeiotic DNA replication), both early and late meiotic genes are underexpressed relative to wild type at late meiotic time points as the cells die. Genes that are differentially expressed relative to wild type in both mutants are particularly strong candidates for playing roles in meiosis and sporulation.
Meiosis is a specialized process of cell division in which one round of DNA replication is followed by two divisions to produce four haploid daughter nuclei. Thousands of genes are differentially expressed in meiotic cells compared with nonmeiotic cells, but genes that are specifically induced in meiosis are not necessarily essential for meiosis (Schlecht and Primig 2003). In addition, a large proportion of genes exhibit changing expression during the course of meiosis (Chu et al. 1998; Primig et al. 2000; Mata et al. 2002; Burns et al. 2010). In Saccharomyces cerevisiae meiosis, the timing of gene induction has been shown to correspond to the time of protein function in many cases, for example, in genes involved in recombination, the pachytene checkpoint, and spore wall formation (Schlecht and Primig 2003).
The basidiomycete mushroom Coprinopsis cinerea, also known as Coprinus cinereus (Redhead et al. 2001), is particularly well suited to studies of gene expression in meiosis because meiosis occurs synchronously in basidial cells, facilitating tissue collection at defined meiotic stages (Figure 1). Burns et al. (2010) examined meiotic expression of genes that have orthologs in C. cinerea, S. cerevisiae, and Schizosaccharomyces pombe and found that genes with meiotic function are more likely to be coinduced on entry to meiosis and more likely to have correlated expression patterns during meiosis than genes not known to be meiotic. This is true even though transcription factors that are major regulators of meiosis differ among the three fungi. For example, in S. cerevisiae Ndt80 induces the expression of hundreds of middle meiotic genes (Chu et al. 1998), but C. cinerea and S. pombe lack identifiable orthologs of this transcription factor. Similarly, in S. pombe the forkhead transcription factor Mei4 is required for the expression of middle meiotic genes (Mata et al. 2002), but its ortholog in S. cerevisiae is not meiosis-specific (Pramila et al. 2006). The C. cinerea genome contains predicted orthologs of forkhead transcription factors but the roles of the encoded proteins are not known.
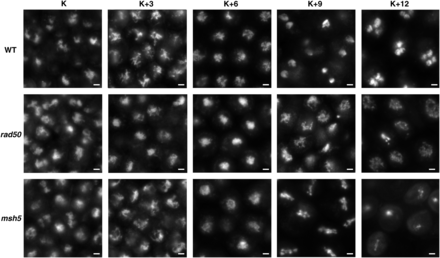
Wild-type, rad50-1, and msh5-22 nuclei during meiosis. Wild-type, rad50-1, and msh5-22 nuclei at the five time points were stained with DAPI, and images were taken at 1000× magnification. Nuclei fuse at karyogamy, condense at K+3, and are fully synapsed at K+6. At K+9, wild-type nuclei are undergoing the first meiotic division, rad50-1 nuclei have arrested in either a diffuse or metaphase-like state, and msh5-22 nuclei are arrested at metaphase I. At K+12, wild-type nuclei have undergone the second meiotic division, rad50-1 nuclei are still arrested, and many msh5-22 cells have died. Scale bars are 2 μm.
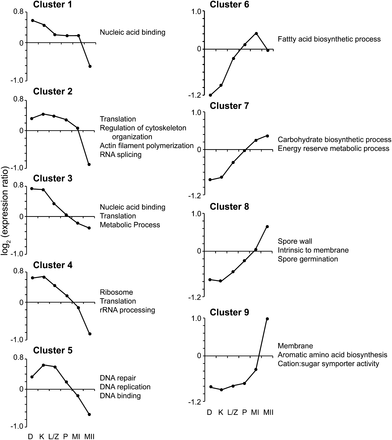
Nearly 3000 of the approximately 13,400 C. cinerea genes (Stajich et al. 2010) change in transcript level during meiosis. These genes are grouped into nine clusters based on their expression patterns over a time course spanning meiosis from before premeiotic DNA replication to after tetrad formation [Figure 2 (Burns et al. 2010)]. Overall, genes in clusters 1 through 5 (early clusters) have relatively high transcript levels at the beginning of meiosis, and these clusters are enriched for genes involved in early meiotic processes with functions such as nucleic acid binding, cytoskeleton organization, chromosome cohesion, and damaged-DNA binding. Genes in clusters 6 through 9 (late clusters) have their greatest transcript abundance at the end of meiosis, and these clusters are enriched for genes involved in gill maturation and sporulation. Mushroom development occurs concurrently with meiosis and spore formation, so only a subset of the genes with changing expression is likely involved in meiosis and sporulation. Therefore, by comparing wild-type gene expression with expression in mutants that do not complete meiosis but have normal mushroom development, we can identify the genes most likely to be responsible for meiosis and sporulation. In this study, we used a mutant that arrests in diplotene (rad50-1) and one that arrests in metaphase I (msh5-22).
Clusters of genes differentially expressed during wild-type meiosis. Line graphs give the average expression profile of each cluster. The Y-axis shows log2-transformed expression ratios. Representative Gene Ontology categories enriched in each cluster are listed. Adopted from Burns et al. (2010).
Mre11, Rad50, and Nbs1 form the highly conserved MRN complex, which is involved in DNA double-strand break (DSB) repair. At the beginning of meiosis, DSBs are induced by the endonuclease Spo11. The MRN complex, along with the nucleases Ctp1 and Exo1, resects the 5′ ends of the DSBs (Nicolette et al. 2010) and the resulting single-strand end is coated with Rad51. This single-strand end then invades the homologous chromosome and subsequent intermediates can be resolved as crossovers. Rad50 is a structural maintenance of chromosomes protein that can dimerize to create a complex of the proper size to bridge sister chromatids or DSB ends (Hopfner et al. 2002). The C. cinerea mutant rad50-1 produces a putative Rad50 protein that is truncated after 360 of the 1309 amino acids of the wild-type protein and is likely unstable (Acharya et al. 2008). S. cerevisiae rad50S-like mutations (certain mutations of RAD50 and null mutations of SAE2) inhibit the resection of DSBs, causing rad50S mutants to delay for several hours in prophase before eventually entering meiosis (reviewed by Hochwagen and Amon 2006). In C. cinerea rad50-1, the number of Rad51 foci is not significantly different from the number of Rad51 foci in a spo11 mutant in which no DSBs are made (Acharya et al. 2008), implying that DSBs are not resected properly in rad50-1. Presumably due to a pathway similar to that causing the arrest of rad50S mutants, 82% of C. cinerea rad50-1 cells arrest in diffuse diplotene and an additional 16% arrest in a metaphase-like state (Figure 1). This mutant has only 2% spore viability, compared with more than 90% in wild type (Acharya et al. 2008).
The C. cinerea msh5-22 mutant fails to replicate DNA before entering meiosis (Kanda et al. 1990). Msh5, a homolog of bacterial MutS, forms a heterodimer with Msh4 that is involved in crossover formation, possibly promoting or stabilizing single-strand invasion intermediates (reviewed by Lynn et al. 2007). Mouse Msh5-deficient spermatocytes and oocytes arrest in zygotene with aberrant chromosome synapsis and then undergo apoptosis (De Vries et al. 1999). Though the absence of premeiotic S phase in msh5 mutants is apparently unique to C. cinerea, cell death is similar; msh5-22 cells die after arresting at metaphase I [Figure 1 (Lu et al. 2003)].
We used microarrays to compare gene expression in rad50-1 and msh5-22 to that in wild type during a 12-hour time course spanning meiosis and identified genes that are differentially expressed at each time point. Our work identified genes whose encoded proteins are strong candidates for playing roles in meiosis and sporulation.
Materials and Methods
Wild-type monokaryon strains J6;5-5 and J6;5-4 (Valentine et al. 1995) and mutant strains rad50-1 (Ramesh and Zolan 1995) and msh5-22 (F. Kennedy, O. Savytskyy, M. Zolan, unpublished data) were used. Mushrooms were grown, RNA was isolated from gill tissue, arrays were hybridized and scanned, spots were flagged, data were normalized, and S. cerevisiae orthologs were identified as previously described (Burns et al. 2010), with the exception that spots on the arrays were not flagged for omission if the percentage of pixels above background plus 1 standard deviation in both channels <60. For each mutant at each time point, RNA was isolated from four biological replicates (each containing seven to ten mushrooms). Samples were collected 3 hr before karyogamy (msh5-22 only), at karyogamy, and 3, 6, 9, and 12 hr after karyogamy. Single layers of gill tissue were stained with DAPI as in Acharya et al. (2008) to confirm that samples were at the correct stage. Each biological replicate was hybridized to a separate microarray with a reference pool of 10 wild-type biological replicates from the same time point. Significance Analysis of Microarrays software was used to identify differentially expressed genes at each time point as well as genes that have patterns of expression across the time course that are significantly different from wild type. We used a false-discovery rate (FDR) cutoff of 5%.
Results
Two meiotic mutants have different overall patterns of differential expression in comparison with wild-type expression
We collected meiotic tissue from rad50-1 and msh5-22 mushrooms at five time points spanning meiosis. In C. cinerea, two haploid nuclei fuse (karyogamy) at the beginning of meiosis, and time points were measured in hours from karyogamy (K). Gill tissue was collected at K, K+3 (leptotene/zygotene), K+6 (pachytene), K+9 (first meiotic division), and K+12 (after tetrads have formed) as well as at K-3 for msh5-22 only (Figure 1). For both rad50-1 and msh5-22, microarrays were used to compare gene expression in four separate biological replicates at each time point to that of a wild-type reference pool of the same time point. We identified genes that are significantly differentially expressed compared to wild type in each mutant with an FDR cutoff of 5%. This means that 5% of the genes included in a list of differentially expressed genes are expected to be false positives.
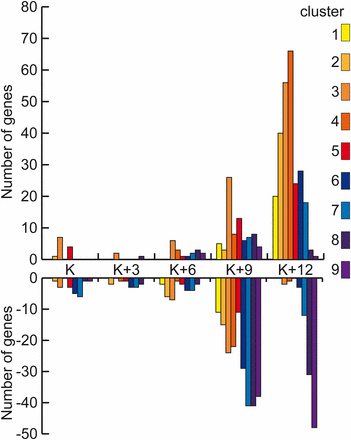
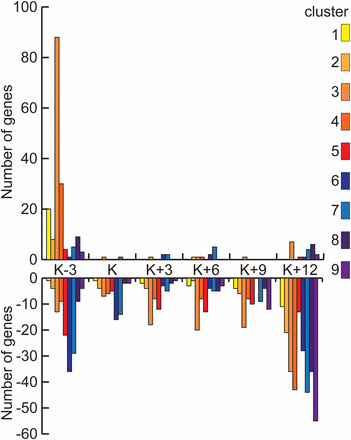
In rad50-1, dozens of genes are differentially expressed at the first three time points, increasing to roughly 1000 by K+12 (FDR 5%; Table 1). To determine whether changes in gene expression reflect the rad50-1 prophase arrest phenotype, we categorized the genes that are differentially expressed in rad50-1 according to their cluster in the wild type. As expected, genes that are overexpressed in rad50-1 compared with the wild type at the end of meiosis tended to be in early clusters whereas those that are underexpressed compared with the wild type at K+12 tended to be in late clusters (Figure 3). Of the genes that are differentially expressed in rad50-1 at K+12 and are in clusters, 91% of those that had greater transcript abundance than wild type were in clusters 1 through 6, whereas 94% of those that are lower than wild type were in clusters 7 through 9. This finding suggests that the expression of a group of genes whose wild-type expression decreases remains high in rad50-1. Conversely, a group of genes that are induced at the end of wild-type meiosis fails to be transcribed in rad50-1. This pattern is consistent with our previous observation that rad50-1 cells arrest in late prophase I (Ramesh and Zolan 1995; Acharya et al. 2008).
Number of differentially expressed genes in rad50-1
| . | K . | K+3 . | K+6 . | K+9 . | K+12 . |
|---|---|---|---|---|---|
| Greater in rad50-1 | 18 | 9 | 45 | 236 | 726 |
| Lower in rad50-1 | 38 | 30 | 94 | 621 | 209 |
| . | K . | K+3 . | K+6 . | K+9 . | K+12 . |
|---|---|---|---|---|---|
| Greater in rad50-1 | 18 | 9 | 45 | 236 | 726 |
| Lower in rad50-1 | 38 | 30 | 94 | 621 | 209 |
The numbers of genes whose transcripts are more and less abundant in rad50-1 at FDR 5% than in wild type at each of five time points are shown. Expression ratios and FDRs for all genes are given in Table S3. FDR, false-discovery rate.
| . | K . | K+3 . | K+6 . | K+9 . | K+12 . |
|---|---|---|---|---|---|
| Greater in rad50-1 | 18 | 9 | 45 | 236 | 726 |
| Lower in rad50-1 | 38 | 30 | 94 | 621 | 209 |
| . | K . | K+3 . | K+6 . | K+9 . | K+12 . |
|---|---|---|---|---|---|
| Greater in rad50-1 | 18 | 9 | 45 | 236 | 726 |
| Lower in rad50-1 | 38 | 30 | 94 | 621 | 209 |
The numbers of genes whose transcripts are more and less abundant in rad50-1 at FDR 5% than in wild type at each of five time points are shown. Expression ratios and FDRs for all genes are given in Table S3. FDR, false-discovery rate.
Number of differentially expressed genes in rad50-1 by expression cluster. At each time point, the number of genes in each cluster whose transcripts are significantly more and less abundant in rad50-1 than in the wild type is shown. Clusters 1−5, which contain genes that are expressed at greater levels at the beginning of meiosis, are represented with warm colors, and clusters 6−9, which contain genes that are expressed at greater levels at the end of meiosis, are represented with cool colors.
We hypothesized that in arrested rad50-1 cells, genes involved in cell-cycle progression would be differentially expressed compared with the wild type. Indeed, of the 12 C. cinerea genes whose S. cerevisiae orthologs are involved in regulation of cyclin-dependent kinase activity, six are differentially expressed at one or both of the last two time points. The most striking example is the single C. cinerea homolog of the S. cerevisiae cyclin genes PCL1, 2, and 9 [CC1G_09963]. In the wild type C. cinerea, transcript abundance of this gene increases more than 10-fold from the beginning to the end of meiosis. The gene has significantly lower transcript abundance in rad50-1 than in wild type at K+9 and K+12. Interestingly, it also is underexpressed in msh5-22 at the same time points. Taken together, this evidence suggests that transcription of this gene increases dramatically throughout meiosis but that this increase is halted at K+9 when the mutants arrest.
Other cell-cycle-related genes have elevated expression in rad50-1 compared with the wild type at one or both of the last two time points (supporting information, Table S1). These include genes that encode the orthologs of S. cerevisiae B-type cyclins (Clb1-4) [CC1G_06397, CC1G_05320] and the catalytic subunit of the main cyclin-dependent kinase (Cdc28) [CC1G_02703], as well as Cdc20 [CC1G_12115], the activator of the anaphase-promoting complex (APC), and an APC component (Apc2) [CC1G_02840]. Thus, control of the checkpoint activated by aberrant repair of meiotic DSBs is likely mediated, at least in part, by transcriptional regulation of cyclins, Cdks, and APC components.
Gene expression in msh5-22 follows a generally different pattern from that seen in rad50-1, particularly at the beginning of the time course. At K-3, 390 genes are expressed below wild-type levels, and 479 genes, 88 of which are in cluster 3, have above wild-type expression levels (Figure 4). Among the genes with higher-than-wild-type transcript levels are several that encode orthologs of budding yeast proteins involved in DNA replication and repair (Table S2): Cdc7 [CC1G_08833], which is required for firing of origins of DNA replication and replication fork progression (Bousset and Diffley 1998); Mcm7 [CC1G_01238], which is part of a complex that promotes DNA melting and elongation during S phase (Tye 1999); Pri2 [CC1G_11942], a component of DNA primase (Foiani et al. 1989); and Rfa2 [CC1G_13904], which is a subunit of the single-stranded DNA binding protein RPA that is involved in DNA replication, repair, and recombination (Longhese et al. 1994). rad50 transcript levels also are greater in msh5-22 than in the wild type at K-3. Genes encoding the ribonucleotide reductase complex (RNR1-4) are transcriptionally induced in response to DNA damage and replication blocks in S. cerevisiae (Huang et al. 1998). This complex catalyzes the rate-limiting step of deoxyribonucleotide biosynthesis and is required for DNA replication and repair (Elledge et al. 1993). In C. cinerea, the ortholog of RNR1 and RNR3 [CC1G_03383] and the ortholog of RNR2 [CC1G_00201] have increased expression in msh5-22 at K-3. These examples suggest that by K-3, msh5-22 cells have already encountered problems in DNA replication and in response have up-regulated transcription (or down-regulated mRNA degradation) of certain DNA replication and repair genes.
Number of differentially expressed genes in msh5-22 by cluster. At each time point, the number of genes in each cluster whose transcripts in msh5-22 are significantly more and less abundant than in the wild type is shown. Warm-colored bars represent clusters 1−5, which contain genes whose expression is greatest at the beginning of meiosis, and cool-colored bars represent clusters 6−9, which contain genes whose expression is greatest at the end of meiosis.
From K through K+9, the number of genes differentially expressed in msh5-22 is fairly constant, with fewer than 40 higher in msh5-22 compared with the wild type and roughly 150 lower (FDR 5%; Table 2). At K+12, the number of differentially expressed genes increases to 75 with greater and 773 with lower expression than wild type. At this point, genes from all nine clusters are underexpressed, as opposed to in rad50-1 in which nearly all the genes with low transcript abundance compared with the wild type are in late clusters (Figure 4). Genes that are expressed at lower-than-wild-type levels in msh5-22 at K+12 include some presumably involved in cell-cycle progression (e.g., homologs of S. cerevisiae CDC53 [CC1G_10644] and SWE1 [CC1G_01961]), DNA replication (e.g., homolog of S. cerevisiae DBF4 [CC1G_07408]), and DNA repair (e.g., rad51 [CC1G_10538]).
Number of differentially expressed genes in msh5-22
| . | K-3 . | K . | K+3 . | K+6 . | K+9 . | K+12 . |
|---|---|---|---|---|---|---|
| Greater in rad50-1 | 479 | 9 | 19 | 39 | 2 | 75 |
| Lower in rad50-1 | 390 | 135 | 146 | 170 | 152 | 773 |
| . | K-3 . | K . | K+3 . | K+6 . | K+9 . | K+12 . |
|---|---|---|---|---|---|---|
| Greater in rad50-1 | 479 | 9 | 19 | 39 | 2 | 75 |
| Lower in rad50-1 | 390 | 135 | 146 | 170 | 152 | 773 |
The numbers of genes whose transcripts are more and less abundant in msh5-22 at FDR 5% than in the wild type at each of six time points are shown. Expression ratios and FDRs for all genes are given in Table S4. FDR, false-discovery rate.
| . | K-3 . | K . | K+3 . | K+6 . | K+9 . | K+12 . |
|---|---|---|---|---|---|---|
| Greater in rad50-1 | 479 | 9 | 19 | 39 | 2 | 75 |
| Lower in rad50-1 | 390 | 135 | 146 | 170 | 152 | 773 |
| . | K-3 . | K . | K+3 . | K+6 . | K+9 . | K+12 . |
|---|---|---|---|---|---|---|
| Greater in rad50-1 | 479 | 9 | 19 | 39 | 2 | 75 |
| Lower in rad50-1 | 390 | 135 | 146 | 170 | 152 | 773 |
The numbers of genes whose transcripts are more and less abundant in msh5-22 at FDR 5% than in the wild type at each of six time points are shown. Expression ratios and FDRs for all genes are given in Table S4. FDR, false-discovery rate.
rad50-1 and msh5-22 do not prepare for postmeiotic DNA replication
After wild-type meiosis, the daughter nuclei migrate into four separate spores and then undergo mitosis to produce binucleate spores (Lu and Jeng 1975; Kamada et al. 1976). The postmeiotic DNA replication that occurs shortly before or after nuclei migrate into the spores cannot take place in arrested rad50-1 and msh5-22 cells, and, as expected, this is reflected in gene expression differences (Table S1 and Table S2). The transcript of the prereplicative complex component protein Cdc6 [CC1G_03323] is less abundant in both mutants at K+9 than in the wild type. Also, pri1 [CC1G_07347] and pri2 [CC1G_11942] transcripts (genes encoding subunits of DNA primase) are less abundant at K+9 in rad50-1, and pri2 is differentially expressed over the time course in msh5-22. Furthermore, expression of the core histones (H2A [CC1G_03522, CC1G_03575, CC1G_07640], H2B [CC1G_03523, CC1G_07639], H3 [CC1G_04396, CC1G_05766, CC1G_08799], and H4 [CC1G_05458, CC1G_05756, CC1G_05765]) as well as the linker histone (H1 [CC1G_03810, CC1G_03813, CC1G_13972]) is lower than in the wild type at K+9 in both mutants.
There seem to be selective pressures that favor both confining core histone expression to S phase and maintaining histone genes in large clusters (reviewed by Marzluff and Duronio 2002). This results in the coordinate expression of histone genes, ensuring that at the time of DNA synthesis, the correct stoichiometric amount of each core histone is produced for proper nucleosome assembly (reviewed by Marzluff and Duronio 2002). Both of these trends seen in other organisms hold true in C. cinerea. For example, there are two pairs of adjacent H2A and H2B genes with the pairs located within 350 kb of each other on chromosome 9 (Broad Institute of Harvard and MIT).
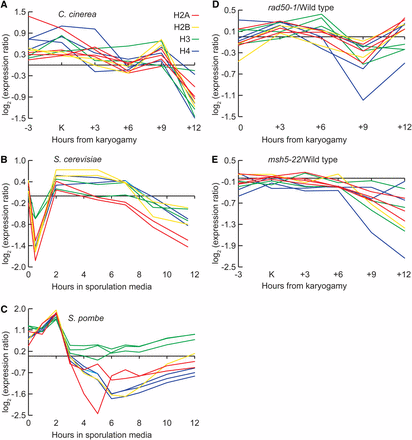
We asked how histone expression in C. cinerea meiosis compares with that in yeasts. In S. cerevisiae, premeiotic DNA replication occurs at roughly 2 hr after transfer to a sporulation-inducing medium. Histone expression increases sharply at this time compared with 1.5 hr earlier and then decreases over the rest of meiosis [Figure 5B (Chu et al. 1998)]. In S. pombe, histone expression also peaks at the time of replication [2 hr (Mata et al. 2002) Figure 5C]. Similarly, we found that core histone transcript abundance in wild-type C. cinerea is high around karyogamy (the approximate time of premeiotic DNA replication) and subsequently decreases. Unlike in the yeasts (in which DNA is not replicated immediately after meiosis) histone expression peaks again at K+9 (Figure 5A) in preparation for postmeiotic S phase. This does not occur in rad50-1 and msh5-22 (Figure 5, D and E), which is consistent with our hypothesis that the cells are not preparing for postmeiotic DNA replication.
Histone expression during meiosis in C. cinerea, S. cerevisiae, and S. pombe. Histone transcript levels for each core histone gene are shown for (A) C. cinerea, (B) S. cerevisiae (data from Chu et al. 1998), and (C) S. pombe (data from Mata et al. 2002). Red lines represent H2A, yellow H2B, green H3, and blue H4. There are two to four copies of each histone gene in the C. cinera genome, and each line represents the transcript levels of one copy. The ratio of transcript abundance in rad50-1 (D) or msh5-22 (E) to the wild type is shown at each time point.
Genes that fail to be induced in both mutants are likely involved in sporulation
For each mutant, we also asked which genes have different overall patterns of expression compared with the wild type across the time course. This type of analysis identified genes for which the expression ratio compared with the wild type changes over the time course. In contrast, the time-point analysis generated a list of the differentially expressed genes at each separate time point (Table S1 and Table S2). For example, a gene that was expressed at one half the level of wild type for the entire time course would be found only by the individual time-point analysis, whereas a gene that was initially expressed at a higher level than wild type and later at a lower level would be recognized by the time course analysis. With a cutoff of 5% FDR, there are 115 genes that have changing expression ratios over the time-course in rad50-1 (Table S1) and 749 in msh5-22 (Table S2; the msh5-22 data include an additional time point (K-3), so these numbers are not directly comparable). These genes include some that were not significantly differentially expressed at any of the individual time points (perhaps because the change at each time point was very small), e.g., those encoding APC components Cdc27 [CC1G_03915] in msh5-22 and Apc1 [CC1G_09696] in rad50-1.
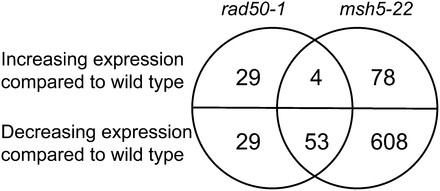
We next asked how much overlap there is between the genes that are differentially expressed over the time course in rad50-1 and those that are differentially expressed in msh5-22. Four of the same genes are overexpressed compared with the wild type over the time course in the two mutants, and 53 genes are underexpressed in both mutants (FDR 5%; Figure 6). Of the 57 genes that are differentially expressed over the time course in both mutants, 60% are meiotically induced in the wild type, which is significantly more than the percentage of all genes (35%) that are meiotically induced (P < 0.0001, χ2 test). This supports our prediction that genes that are differentially expressed in both mutants are likely to be involved in meiosis.
Overlap between genes that are differentially expressed over the time course in rad50-1 and msh5-22. Based on the time course analysis, the numbers of genes whose transcripts change significantly in each mutant with respect to the wild type are shown. Four genes have increasing transcript abundance, and 53 have decreasing transcript abundance compared with the wild type in the two mutants.
We also predicted that genes that are differentially expressed in both mutants at late time points are likely to be involved in sporulation. Twenty-four of the genes that are differentially expressed over the time course in both mutants have changing expression during wild-type meiosis. Of these, 17 are in cluster 9 (which is enriched for genes involved in spore formation) and four more are in other late clusters. For example, the ortholog of the S. cerevisiae gene ARO2 [CC1G_03718] was identified by the time-course analysis as having decreasing transcript abundance with respect to wild type over the time-course in both mutants. In S. cerevisiae, this protein participates in aromatic amino acid synthesis and is required for sporulation (Lucchini et al. 1978). Sporulation requires the biogenesis of the multilayered spore wall (Kues 2000), and the genes that are differentially expressed in both mutants include orthologs of S. cerevisiae genes involved in this process. For example, Exg1 [CC1G_06563] is involved in cell wall β-glucan assembly (Vazquez De Aldana et al. 1991). Other genes putatively involved in posttranslational processing of extracellular proteins are also differentially expressed: in S. cerevisiae, Pdi1[CC1G_00344] is essential for formation of disulfide bonds in secretory and cell-surface proteins (Freedman 1989) and Pmt4 [CC1G_00834] is involved in O-mannosylation of secretory proteins (Strahl-Bolsinger et al. 1993).
At K+12, there are 120 genes whose expression is lower than that in the wild type in both mutants, and one-half of these are in late clusters in the wild type, including 31 in cluster 9. For example, the orthologs of the S. cerevisiae genes ARO1 [CC1G_09809], ARO2, and the sporulation-specific chitinase gene CTS2 [CC1G_04870] (Dunkler et al. 2008) are all in cluster 9 in the wild type. Orthologs of other S. cerevisiae genes encoding proteins required for cell wall synthesis and assembly, e.g., GAS1 [CC1G_13682] (Popolo et al. 1993) and ROT2 [CC1G_04516] (Trombetta et al. 1996), which are not in any cluster in wild type, are also underexpressed in both mutants. This supports the hypothesis that genes differentially expressed at the end of the time course in both mutants are likely to play roles in sporulation. Also, some of the genes that this analysis indicates are likely to be involved in sporulation were not in wild-type clusters. This means that identifying genes not induced at the end of meiosis in the mutants and identifying genes that are up-regulated during sporulation in wild type are complementary methods of learning which genes are involved in sporulation. Genes in clusters 6 and 7 play roles in gill expansion (which happens normally in the mutants), whereas clusters 8 and 9 are enriched for genes involved in spore structure and packaging and preparation for spore germination (Burns et al. 2010). Spores are not made in the mutants, so it is not surprising that roughly one-third of the genes that are differentially expressed in both mutants are in cluster 9.
Discussion
Identification of new meiotic and sporulation genes
Nearly 3000 genes are differentially expressed during the synchronous meiosis and mushroom development of C. cinerea (Burns et al. 2010). To determine which of these are involved in meiosis itself, we identified genes whose expression is significantly different from the wild type in two mutants that arrest in meiosis but otherwise produce morphologically normal mushrooms. In both cases, the number of differentially expressed genes increases dramatically beginning at the time point when the mutant arrests. The lists of differentially expressed genes include some whose S. cerevisiae orthologs are known to encode proteins with roles in cell cycle progression, S phase, and sporulation. This supports our prediction that genes differentially expressed in the mutants are indeed likely to be involved in meiotic processes. In addition to identifying genes involved in wild-type meiosis, this study contributes to an understanding of what happens when errors arise in meiosis and how changes in transcription mediate and result from checkpoint arrests. Checkpoint activation leads to transcriptional changes and these changes then effect cell cycle arrest and the observed morphological changes in the mutants, e.g. lack of spore formation.
rad50-1 and msh5-22 show checkpoint arrest
Several different checkpoint pathways can be activated in response to errors in meiosis. Each checkpoint consists of a signal, which is detected by signal sensors that activate signal transduction pathways, which in turn modify targets to produce an output. Various errors in DSB processing create signals (e.g., Rad51-coated single-stranded DNA), and through checkpoint pathways, these signals lead to the modification of targets with the result of controlling cell-cycle progression, DNA repair, programmed cell death, and in some cases development (reviewed by Hochwagen and Amon 2006). Mutations in rad50 and msh5 trigger checkpoints in other organisms (De Vries et al. 1999; Usui et al. 2001; Borner et al. 2004), rad50-1 and msh5-22 each exhibit errors in DSB processing in C. cinerea [(Acharya et al. 2008) F. Kennedy, O. Savytskyy, M. Zolan, unpublished data], and our microscopy observations indicate that meiosis stalls before the first division in both mutants (Figure 1). Moreover, our expression data show that in rad50-1 cells, a group of early meiotic genes continue to have high transcript levels through K+12, whereas conversely the expression of dozens of genes that are expressed at the end of meiosis in wild type is never triggered. Thus, we believe that meiotic checkpoint pathways are being activated in the two mutants studied. We have shown that the arrest also prevents the induction of some genes involved in spore formation and postmeiotic DNA replication, and in rad50-1 prevents the down-regulation of many prophase genes. Our results highlight the overlap between presumed checkpoint targets in these two mutants, which have different arrest phenotypes and presumably different checkpoint signals.
rad50-1 gene expression reflects mutant phenotypes
Few genes are differentially expressed in rad50-1 compared with the wild type at the beginning of the time course (Table 1) meaning that the loss of Rad50 function has little effect on early expression of meiotic genes. This finding is consistent with the observation that rad50-1 chromosomes condense, pair, and synapse with the same timing as wild type, though at much lower levels [Figure 1 (Acharya et al. 2008)]. Kugou et al. (2007) explored the effect of MRN-complex mutations on meiotic gene expression in S. cerevisiae. They compared gene expression at the time of transfer to sporulating media to expression 4 hr later in both wild-type and an mre11-null mutant. At this time point [which corresponds to K+3 in C. cinerea (Burns et al. 2010)], many key meiotic genes were similarly regulated in mutant and wild-type cells, but the expression of a group of genes enriched for spore wall biogenesis functions depended on Mre11. At the same time in S. cerevisiae rad50Δ, the overall induction of meiotic genes was largely unaffected (Kugou et al. 2007). This evidence suggests that MRN mutations may affect meiotic gene expression in S. cerevisiae, but because the study examined only a single early time point, it does not give a complete picture of the expression changes.
Checkpoint activation in msh5-22
Unlike rad50-1, all msh5-22 cells progress to metaphase, arrest, and then at roughly K+10 start to die [Figure 1 (Lu et al. 2003)]. In msh5-22, DNA is not replicated before meiosis, raising the possibility that a DSB checkpoint similar to the mechanism in S. cerevisiae and S. pombe is triggered. In the yeasts, this checkpoint ensures that DSBs are not formed in a region until the replication fork has passed that location (Borde et al. 2000; Tonami et al. 2005). This mechanism is not activated unless DNA replication has been initiated (Murakami and Nurse 2001), raising the alternative possibility that a DSB checkpoint exists in C. cinerea but is bypassed in msh5-22 if DNA replication is never initiated. The number of Rad51 foci (which mark resected DSBs) in msh5-22 is the same as the number of foci in a mutant (spo11-1) that fails to make DSBs, implying that meiotic DSBs are not induced in msh5-22 (F. Kennedy, O. Savytskyy, M. Zolan, unpublished data). The apparently extremely low level of DSBs is consistent with the hypothesis that a checkpoint preventing their formation is triggered in this mutant.
In rad50-1, there is a large group of early genes whose transcript abundance remains high at late time points; however, this pattern is not evident in msh5-22, likely because msh5-22 cells do not arrest until metaphase I, which occurs just before K+9. By K+12, the end of the time course, many msh5-22 cells have already died, which explains why genes from all clusters have low transcript abundance at this point. In wild-type cells at metaphase I, tension is established though crossovers between homologous chromosomes, cohesion between sister chromatids, and amphitelic attachment of homologs. msh5-22 chromosomes have very few crossovers and lack sister chromatids (F. Kennedy, O. Savytskyy, M. Zolan, unpublished data). The absence of tension resulting from these defects could trigger the spindle assembly checkpoint, leading to the observed metaphase arrest.
Similarities between rad50-1 and msh5-22
In C. cinerea, genes that are coregulated with meiotic genes are expected to have meiotic roles, but genes involved in other processes, such as mushroom development, also are likely to be induced during meiosis. Because mushroom development proceeds normally in both rad50-1 and msh5-22, genes that are not differentially expressed in the mutants compared with the wild type are not likely to be involved in meiosis or spore formation. The 206 genes that are in early clusters in wild type (Burns et al. 2010) and also have high expression at K+12 in rad50-1 are those most likely to play interesting roles in early meiotic processes. Although genes in early clusters that are differentially expressed in the mutants at the end of meiosis tend to have elevated expression compared with the wild type, there is a group of 83 genes in early clusters that are underexpressed at K+9 in rad50-1, including the histones. Histones have high expression early in meiosis (Burns et al. 2010) but their synthesis is also necessary for post-meiotic DNA replication, suggesting that other genes in this category could similarly play both early and late roles. The 120 genes that are underexpressed in both rad50-1 and msh5-22 at K+12 are the strongest candidates for involvement in sporulation.
Recently, Nakazawa et al. (2011) created a C. cinerea ku70 mutant and showed that in the absence of this non-homologous end joining protein, an exogenous copy of a gene introduced by transformation disrupts the genomic copy at a high frequency. When this strain is used, it would be feasible to knock out the candidate genes identified in our study. Alternatively, RNAi techniques are available to knock down gene expression in C. cinerea (Namekawa et al. 2005; Walti et al. 2006). Therefore, it would be straightforward knock out or knock down gene function to test the roles of candidate genes in the processes of meiosis and sporulation.
Acknowledgments
We thank the Indiana University Center for Genomics and Bioinformatics, particularly John Colbourne and Jackie Lopez, for providing microarray assistance, scanning equipment, and software. We are grateful to Sasha Savytskyy for microscopy assistance and Hong Fan for general lab support. This research was funded by National Institutes of Health grant GM43930 (to M.E.Z.), the Indiana METACyt Initiative of Indiana University (which is funded in part through a major grant from the Lilly Endowment, Inc.), and the Beckman Foundation.
Literature Cited
Broad Institute of Harvard and MIT. Coprinopsis cinerea Sequencing Project. Available at: http://www.broadinstitute.org/.
Footnotes
Communicating editor: M. S. Sachs
Author notes
This is an open-access article distributed under the terms of the Creative Commons Attribution Unported License (http://creativecommons.org/licenses/by/3.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.112.003046/-/DC1
Raw microarray data from this article have been deposited in the GEO database under accession no. GSE37968 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE37968).
Present address: Washington & Jefferson College, 60 South Lincoln Street, Washington, PA 15301.