-
PDF
- Split View
-
Views
-
Cite
Cite
Ronda L Hamm, Richard P Meisel, Jeffrey G Scott, The Evolving Puzzle of Autosomal Versus Y-linked Male Determination in Musca domestica, G3 Genes|Genomes|Genetics, Volume 5, Issue 3, 1 March 2015, Pages 371–384, https://doi.org/10.1534/g3.114.014795
Close - Share Icon Share
Abstract
Sex determination is one of the most rapidly evolving developmental pathways, but the factors responsible for this fast evolution are not well resolved. The house fly, Musca domestica, is an ideal model for studying sex determination because house fly sex determination is polygenic and varies considerably between populations. Male house flies possess a male-determining locus, the M factor, which can be located on the Y or X chromosome or any of the five autosomes. There can be a single M or multiple M factors present in an individual male, in heterozygous or homozygous condition. Males with multiple copies of M skew the sex ratio toward the production of males. Potentially in response to these male-biased sex ratios, an allele of the gene transformer, Md-traD, promotes female development in the presence of one or multiple M factors. There have been many studies to determine the linkage and frequency of these male determining factors and the frequency of Md-traD chromosomes in populations from around the world. This review provides a summary of the information available to date regarding the patterns of distribution of autosomal, X-linked and Y-linked M factors, the relative frequencies of the linkage of M, the changes in frequencies found in field populations, and the fitness of males with autosomal M factors vs. Y-linked M. We evaluate this natural variation in the house fly sex determination pathway in light of models of the evolution of sex determination.
Sex determination is the initiation of a gene regulatory cascade responsible for the differential expression of genes between males and females, giving rise to reproductive traits and sexually dimorphic phenotypes. Paradoxically, even though sex determination is an essential developmental pathway required for fertility, sex determination pathways evolve extremely fast (Bull 1983; Marin and Baker 1998; Haag and Doty 2005). The genes or environmental cues responsible for the initiation of sex determination (master regulators) often differ between closely related species (Wilkins 1995; Graham et al. 2003). This evolutionary turnover in the initiation of sex determination pathways is contrasted by the use of conserved downstream components across distantly related taxa. For example, genes from the doublesex/mab-3 related (Dmrt) family are involved in sex determination pathways in vertebrates, insects, and round worms (Raymond et al. 1998; Haag and Doty 2005).
Multiple hypotheses have been put forth to explain the evolutionary turnover at the top of sex determination pathways. In one set of models, it was demonstrated that a novel sex determiner can invade a population if it is genetically linked to a beneficial allele (Charlesworth and Charlesworth 1980; Rice 1986). If the allele linked to the sex determiner confers a fitness benefit to one sex and is detrimental to the other sex (i.e., it has a sexually antagonistic fitness effect), the new sex determiner is particularly likely to invade because it resolves the intersexual conflict by limiting the inheritance of the sexually antagonistic allele to the sex in which it is beneficial (Van Doorn and Kirkpatrick 2007, 2010). In another set of models, it was shown that a new sex-determining locus can invade a population if the sex ratio (relative number of breeding males and females) deviates from the equilibrium (often 1:1) (Eshel 1975; Bull and Charnov 1977; Bulmer and Bull 1982). In this case, the new sex determining locus produces a balanced sex ratio.
The house fly, Musca domestica, is a powerful model system for studying the genetics, molecular biology, and evolution of sex determination. The house fly has one of the most polymorphic sex determination pathways of any animal (Bull 1983; Dübendorfer 2001), and the past two decades have seen substantial advances in our understanding of the molecular regulation of the house fly sex determination pathway (Meise et al. 1998; Dübendorfer 2001; Hediger et al. 2004; Burghardt et al. 2005; Hediger et al. 2010; Meier et al. 2013). We highlight features of this polymorphism, and we describe how experiments on house flies have contributed toward our understanding of the evolution of sex determination. The house fly genome was sequenced recently (Scott et al. 2014a), which opens the door for improved understanding of sex determination in this species. We close with predictions for future insights that can be gleaned from technological advances in genomics and potential applications for control of house flies, which are mechanical vectors of scores of human and animal diseases.
Sex determination in house flies
Conserved dipteran sex determination pathway
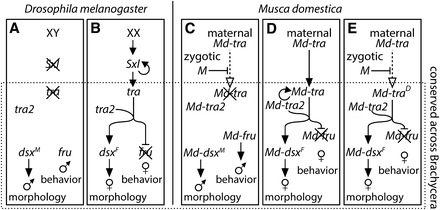
Sex determination in dipterans relies heavily on the differential regulation of alternative splicing between the sexes of genes expressed in both males and females (Salz 2011). The dipteran sex determination pathway (at least in Brachycera, or “higher” dipterans) consists of a core series of regulatory steps that are conserved in all brachyceran species examined thus far (Pomiankowski et al. 2004; Bopp et al. 2014; Geuverink and Beukeboom 2014) (Figure 1). At the start of this core pathway, the splicing regulator transformer (tra) is itself alternatively spliced to produce a functional transcript capable of encoding a full-length protein in females and a nonfunctional transcript with a premature stop codon in males (Boggs et al. 1987; McKeown et al. 1987; Pane et al. 2002, 2005; Lagos et al. 2007; Ruiz et al. 2007; Concha and Scott 2009). The factor responsible for the decision whether to produce the male or female splice form of tra, however, varies across species, as described in the next section.
Sex-determination pathways. The (A) male and (B) female Drosophila sex-determination pathways are shown, along with the house fly (C) male-determining pathway, (D) canonical female-determining pathway, and (E) female-determining pathway via the action of Md-traD. The core of the pathway that is conserved across brachyceran flies is contained within the dashed box. Abbreviations are described in the main text.
Functional TRA protein in females, along with the product of the constitutively expressed transformer 2 (tra2), promotes the splicing of the Dmrt homolog doublesex (dsx) into its female-specific isoform (dsxF), initiating female morphological development (Hoshijima et al. 1991) (Figure 1). TRA also causes the male-specific behavioral regulator fruitless (fru) to be spliced into a nonfunctional isoform in females (Ito et al. 1996; Ryner et al. 1996; Demir and Dickson 2005; Meier et al. 2013). The absence of functional TRA in males leads to male-specific splicing of dsx (dsmM) and splicing of fru into its functional male-specific isoform, initiating the development of male morphology and behavior, respectively.
Variation in sex determination across dipterans
Although the aforementioned core sex determination pathway is conserved among dipterans, there is variation across species in how the pathway is initiated (Bopp et al. 2014). This is consistent with a model whereby sex determination pathways evolve by the change or addition of upstream components, because changes at the top of pathways are less likely to have deleterious effects (Wilkins 1995; Marin and Baker 1998). In the well-studied Drosophila system, tra splicing ultimately is controlled by the number of X chromosomes in the zygote (Bridges 1921; Pomiankowski et al. 2004; Erickson and Quintero 2007; Salz and Erickson 2010). Female zygotes (XX) have greater expression of X-linked “numerator” genes than male zygotes (XY). Two doses of the X-linked numerators leads to the expression of functional Sex lethal (Sxl) transcripts in females (Cline 1988; Duffy and Gergen 1991; Sefton et al. 2000), and the SXL protein autoregulates the continued splicing of Sxl into a functional transcript in females (Cline 1984) (Figure 1B). Functional SXL in females promotes splicing of tra into a functional isoform, whereas lack of functional SXL in males leads to nonfunctional splicing of tra (Valcárcel et al. 1993) (Figure 1A). Sxl is expressed equally in both sexes in other dipterans and is not a master regulator of sex determination in non-Drosophila species (Marin and Baker 1998; Schütt and Nothiger 2000; Saccone et al. 2002; Shearman 2002).
Other mechanisms of initiating the sex determination pathway in dipterans include environmental sex determination (i.e., Aedes stimulans) (Horsfall and Anderson 1963), female-determining factors (i.e., M. domestica and Tephritidae), or maternal genotype (i.e., Sciara and Chrysomya) (Marin and Baker 1998; Saccone et al. 2002). Many dipteran species, including house fly, have a dominant male-determining factor (M) that is thought to inhibit the splicing of tra into a functional isoform in developing male zygotes (Traut and Willhoeft 1990; Dübendorfer et al. 2002; Bopp et al. 2014) (Figure 1C). In house flies, the homolog of tra (Md-tra) is expressed in the maternal germline, and lack of M in female zygotes allows maternal Md-tra to feed forward into zygotic expression of functional Md-tra (Hilfiker-Kleiner et al. 1994; Dübendorfer and Hediger 1998; Bopp 2010; Hediger et al. 2010) (Figure 1D). Zygotic TRA, along with TRA2, autoregulates the continued splicing of Md-tra into a functional isoform in the female zygote, whereas M breaks the feed-forward regulation of Md-tra in male zygotes (Figure 1, C and D) (Bopp 2010).
The ancestral brachyceran sex determination mechanism is hypothesized to be a male-determining gene located on the heteromorphic Y chromosome or one of the homomorphic chromosomes (Saccone et al. 2002; Vicoso and Bachtrog 2013). However, the position of M is not static in some species. In Megaselia scalaris the male-determining factor can be located on the first, second, or third chromosome (Traut 1994), although the transposing nature of the M. scalaris male-determining factor recently has been called into question (Hoehn and Noor 2015). In the mosquito Culex tritaeniorhynchus sex is determined by a male factor located on either linkage group I or III depending on the population (Baker and Sakai 1976). Variation in the linkage of M in house flies is detailed below.
The aforementioned developmental pathway regulates sex determination in somatic tissues. Germline sex determination in dipterans relies upon input from the somatic pathway, but the interdependence of the germline and somatic sex determination pathways is taxon-specific. For example, the Drosophila germline sex determination pathway combines information from the germline genotype with signals from the surrounding soma to determine the sex-specific developmental fate of germline tissues (Defalco et al. 2008; Casper and Van Doren 2009). Germline sex determination in house flies, on the other hand, depends entirely on the genetic sex of the surrounding soma (Hilfiker-Kleiner et al. 1994).
M. domestica: M and F factors
Since the early report by Stevens (1908), many investigators have confirmed that the diploid chromosome number in the standard house fly, M. domestica L., is 12 consisting of 5 pairs of autosomes and a pair of sex chromosomes, and that the male is the heterogametic sex; that is, XX-type for the female and XY-type for the male (Hiroyoshi et al. 1982). The current nomenclature system (Wagoner 1969a) numbers house fly autosomes in order of decreasing length (i.e., autosome I is longest and autosome V is shortest). These “standard” populations are composed of XYM males and XX females. In these populations, maleness is determined by a Y chromosome that harbors a male-determining M factor (YM) (Hiroyoshi 1964; Dübendorfer 2001). Two Y chromosome regions with male-determining activity (i.e., M) have been identified that are functionally equivalent but nonredundant (Hediger et al. 1998). Both Y-linked male-determining regions are required for male development in the absence of autosomal M (AM) or X-linked M (XM) factors (Hediger et al. 1998).
The house fly X and Y chromosomes are largely heterochromatic and lack any known genes aside from M (usually on Y, but occasionally on X, see Table 1) (Bull 1983; Inoue and Hiroyoshi 1986). In addition, the number of Xs or Ys in a karyotype can vary up (i.e., XXY or XXX) or down (i.e., XO, OY) without any effect, as long as one X or Y is present (Bull 1983). Flies carrying only the short arm of the Y chromosome are also viable, but the long arm of the Y is not sufficient for viability in the absence of an X chromosome (Hediger et al. 1998). This suggests that any essential genes on the sex chromosomes must be located on the short arm of the Y (which also has a euchromatic segment) and on the homologous segment of the X chromosome. Almost all other muscid flies have five pairs of euchromatic chromosomes similar to the house fly “autosomes,” but not all species have the heterochromatic pair (Boyes et al. 1964; Bull 1983). However, nearly all species examined in other closely related families have five autosomes and a pair of sex chromosomes (Boyes and Van Brink 1965), suggesting that five autosomes plus the sex chromosomes (2n = 12) is the ancestral karyotype among most calyptrate flies, including muscids.
Percentagesa of M and Md-traD in field collected strains of house fly
| Location . | IM . | IIM . | IIIM . | IVM . | VM . | YMb . | M/+c . | M/Md . | Md-traDe . | Reference . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Africa | S. Africa | Johannesburg-Pretoria area SA1 | 0 | 0 | 100 | 0 | 0 | ✓ | ✓ | (Denholm et al. 1990) | ||
| “ | S. Africa | Johannesburg-Pretoria area SA2 | 0 | 0 | 85 | 0 | 7.4 | 7.4 | 45 | ✓ | “ | |
| “ | S. Africa | Zinkwazi Beach | 0 | ✓ | ✓ | 0 | 0 | 0 | 29 | (Feldmeyer et al. 2008) | ||
| “ | S. Africa | Umhlali | ✓ | ✓ | ✓ | 0 | ✓ | 0 | 79 | “ | ||
| “ | S. Africa | Hammarsdale | 0 | ✓ | ✓ | 0 | 0 | 0 | 92 | “ | ||
| “ | S. Africa | Ashburton | ✓ | ✓ | 0 | 0 | ✓ | 0 | 13 | “ | ||
| “ | S. Africa | Mooi River | 0 | ✓ | ✓ | 0 | 0 | 0 | 29 | “ | ||
| “ | S. Africa | Warden | 0 | 0 | 70 | 0 | 0 | 30 | ✓ | 15 | “ | |
| “ | S. Africa | South Africa combined | 26 | “ | ||||||||
| “ | Tanzania | Same | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Moshi | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Makuiuny | 0 | 80 | 0 | 0 | 0 | 20 | ✓ | 100 | “ | |
| “ | Tanzania | Arusha | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Karatu | 0 | 80 | 0 | 0 | 0 | 20 | ✓ | 85 | “ | |
| “ | Tanzania combined | 62 | “ | |||||||||
| “ | Tanzania | ✓ | (Scott et al. 2014a) | |||||||||
| Australia | Australia | Ipswich | 0 | 44 | 70 | 2 | 0 | 7 | 92 | 70 | ✓ | (Hamm and Scott 2009) |
| “ | “ | Bowhill | 0 | ✓ | ✓ | 0 | ✓ | ✓ | (Wagoner 1969b) | |||
| Asia | Japan | Furano | 0 | 0 | 9 | 0 | 0 | 91 | 0 | 0 | (Tomita and Wada 1989b) | |
| “ | “ | Sapporo | 0 | 0 | 29 | 0 | 0.6 | 70 | 0.3 | 0.6 | “ | |
| “ | “ | Akkeshi | 0 | 0 | 21 | 0 | 0 | 79 | 0 | 0 | “ | |
| “ | “ | Obihiro | 0 | 0 | 12 | 0 | 0 | 88 | 0 | 0 | “ | |
| “ | “ | Hachinohe | 0 | 0 | 38 | 0 | 0.5 | 57 | 2 | 4 | “ | |
| “ | “ | Niharu | 5 | 0 | 32 | 0 | 0 | 64 | 0 | “ | ||
| “ | “ | Togakushi | 0 | 8 | 58 | 0 | 0 | 35 | 4 | 28 | “ | |
| “ | “ | Haga | 0 | 2 | 96 | 0 | 0 | 2 | 4 | 17 | “ | |
| “ | “ | Miyagi | 0 | 3 | 35 | 0 | 0 | 63 | 5 | 0 | “ | |
| “ | “ | Hokota | 0 | 2 | 57 | 0 | 0 | 40 | 5 | “ | ||
| “ | “ | Kofu | 0 | 0 | 70 | 0 | 0 | 30 | 24 | 29 | “ | |
| “ | “ | Yumenoshima | 0 | 1 | 74 | 0 | 0 | 25 | 68 | 48 | 99 | “ |
| “ | “ | Aio | 2 | 31 | 29 | 3 | 22 | 12 | 2 | 1 | “ | |
| “ | “ | Kasuya | 1 | 16 | 39 | 0 | 18 | 26 | 20 | 38 | “ | |
| “ | “ | Nangoku | 3 | 0 | 24 | 0 | 3 | 70 | 6 | 13 | “ | |
| “ | “ | Haruno | 0 | 0 | 34 | 0 | 0 | 66 | 2 | 0 | “ | |
| “ | “ | Hachijo | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | Okinawa | 4 | 41 | 48 | 2 | 0 | 4 | 15 | 47 | “ | |
| “ | “ | Ishigaki | 0 | 32 | 54 | 0 | 0 | 14 | 2 | 4 | “ | |
| “ | “ | Kirishima | 0 | (Hiroyoshi 1964) | ||||||||
| “ | “ | Nichinan | 0 | “ | ||||||||
| “ | “ | Sakurai | 0 | “ | ||||||||
| “ | “ | Kitakyushu | ✓ | ✓ | ✓ | 0 | 0 | ✓* | ✓ | 0 | (Tsukamoto et al. 1980) | |
| “ | “ | Kitakyushu | 0 | ✓ | ✓ | 0 | ✓ | ✓* | ✓ | 0 | “ | |
| “ | “ | FR 83 | 80 | 0 | 0 | (Tomita and Wada 1989a) | ||||||
| “ | “ | OH 83 | 0 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | AK 83 | 0 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-YG 83 | 0 | 0 | 40 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-YG 84 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-OD 84 | 0 | 0 | 33 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | IK-RS 84 | 0 | 0 | 24 | 0 | 0 | 0 | 2 | 3 | “ | |
| “ | “ | IK-YU 84 | 0 | 0 | 30 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | IK-BN 84 | 0 | 0 | 35 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | OT-ZB 84 | 0 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | “ | |
| Osaka | ✓ | “ | ||||||||||
| Asia/Europe | Turkey | Giresun | ✓ | ✓ | (Cakir 1999) | |||||||
| “ | “ | Ordu | ✓ | ✓ | “ | |||||||
| “ | “ | Trabzon | ✓ | ✓ | “ | |||||||
| “ | “ | Giresun | 0 | ✓ | 0 | ✓ | (Cakir and Kence 2000) | |||||
| “ | “ | Trabzon | 0 | 0 | ✓ | ✓ | “ | |||||
| “ | “ | Kayrak | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Simav | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Izmit | ✓ | ✓ | 0 | ✓ | “ | |||||
| “ | “ | Iskenderun | ✓ | ✓ | ✓ | ✓ | “ | |||||
| “ | “ | Balikesirr | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Polatli | 0 | 0 | 0 | “ | ||||||
| Trabzon | ✓ | (Scott et al. 2014a) | ||||||||||
| Europe | British Isles | Fm31 | 0 | 0 | 0 | 0 | 0* | ✓ | ✓ | (Denholm et al. 1985) | ||
| “ | “ | Fm39 | 0 | 0 | ✓ | 0 | ✓ * | ✓ | “ | |||
| “ | “ | Fm42 | 0 | 0 | 0 | 0 | ✓ | “ | ||||
| “ | “ | Harpenden | ✓ | ✓ | ||||||||
| “ | “ | Fm44 | 0 | 0 | ✓ | 0 | ✓ | 25 | 35-52 | “ | ||
| “ | “ | Fm45 | 0 | 0 | ✓ | 0 | ✓ | ✓ | “ | |||
| “ | England | Fm6 | 0 | 0 | 6 | 0 | 0 | 94* | 6 | ✓ | (Denholm et al. 1983) | |
| “ | “ | Fm22 | 0 | 0 | 2.9 | 0 | 0 | 69* | 29 | ✓ | “ | |
| “ | Italy | 12 populations | 0 | 0 | 0 | 0 | 0 | ✓ | (Franco et al. 1982) | |||
| “ | “ | 11 populations | ✓ | ✓ | ✓ | ✓ | “ | |||||
| “ | “ | IT1 | 0 | 0 | 12 | 0 | 0 | 52 | 44 | (Kozielska et al. 2008) | ||
| “ | “ | IT2 | 0 | 25 | 9 | 0 | 0 | 44 | 43 | “ | ||
| “ | “ | IT3 | 0 | 0 | 0 | 0 | 0 | 50 | 10 | “ | ||
| “ | “ | IT4 | 12 | 9 | 45 | 0 | 0 | 42 | ✓ | 100 | “ | |
| “ | “ | IT5 | 2 | 17 | 50 | 0 | 9 | 62 | ✓ | 100 | “ | |
| “ | “ | IT6 | 3 | 13 | 32 | 0 | 0 | 68 | ✓ | 95 | “ | |
| “ | “ | IT7 | 0 | 3 | 53 | 0 | 0 | 17 | 78 | “ | ||
| “ | “ | IT8 | 9 | 3 | 86 | 3 | 3 | 16 | ✓ | 100 | “ | |
| “ | “ | IT9 | 8 | 17 | 46 | 0 | 0 | 6 | 86 | “ | ||
| “ | “ | IT10 | 3 | 0 | 55 | 3 | 0 | 0 | 95 | “ | ||
| “ | “ | IT11 | 0 | 0 | 76 | 0 | 0 | 3 | 96 | “ | ||
| “ | “ | IT12 | 0 | 0 | 56 | 0 | 0 | 8 | 47 | “ | ||
| “ | Switzerland | Switzerland | 0 | 0 | 0 | 0 | 0 | 50 | 5 | “ | ||
| “ | Germany | GE1 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | “ | ||
| “ | “ | GE2 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | “ | ||
| France | Faverges | ✓ | (Scott et al. 2014a) | |||||||||
| Spain | Santa Fé | ✓ | “ | |||||||||
| N. America | USA | Texas | 10 | 0 | ✓ | (McDonald and Overland 1974) | ||||||
| “ | “ | North Dakota | 8 | 0 | ✓ | “ | ||||||
| “ | “ | Florida | 100 | 0 | 0 | “ | ||||||
| “ | “ | Florida | 0 | 0 | 100 | 0 | 0 | 0 | (Hamm et al. 2005) | |||
| “ | “ | North Carolina 2002 | 0 | 0 | 20 | 0 | 0 | 78 | 2.4 | 0 | “ | |
| “ | “ | North Carolina 2006 | 0 | 0 | 19 | 0 | 0 | 78 | 1.4 | 1.4 | (Hamm and Scott 2008) | |
| “ | “ | North Carolina 2007 | 0 | 0 | 2.3 | 0 | 0 | 95 | 0 | 2.3 | 4.2 | “ |
| “ | “ | New York | 0 | 0 | 4.4 | 0 | 0 | 96 | (Hamm et al. 2005) | |||
| “ | “ | Maine | 0 | 0 | 0 | 0 | 0 | 100 | “ | |||
| “ | “ | California- Chino | 0 | 0 | 15 | 0 | 0 | 85 | ✓ | ✓ | (T. Shono and J. G. Scott, personal communication) | |
| Location . | IM . | IIM . | IIIM . | IVM . | VM . | YMb . | M/+c . | M/Md . | Md-traDe . | Reference . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Africa | S. Africa | Johannesburg-Pretoria area SA1 | 0 | 0 | 100 | 0 | 0 | ✓ | ✓ | (Denholm et al. 1990) | ||
| “ | S. Africa | Johannesburg-Pretoria area SA2 | 0 | 0 | 85 | 0 | 7.4 | 7.4 | 45 | ✓ | “ | |
| “ | S. Africa | Zinkwazi Beach | 0 | ✓ | ✓ | 0 | 0 | 0 | 29 | (Feldmeyer et al. 2008) | ||
| “ | S. Africa | Umhlali | ✓ | ✓ | ✓ | 0 | ✓ | 0 | 79 | “ | ||
| “ | S. Africa | Hammarsdale | 0 | ✓ | ✓ | 0 | 0 | 0 | 92 | “ | ||
| “ | S. Africa | Ashburton | ✓ | ✓ | 0 | 0 | ✓ | 0 | 13 | “ | ||
| “ | S. Africa | Mooi River | 0 | ✓ | ✓ | 0 | 0 | 0 | 29 | “ | ||
| “ | S. Africa | Warden | 0 | 0 | 70 | 0 | 0 | 30 | ✓ | 15 | “ | |
| “ | S. Africa | South Africa combined | 26 | “ | ||||||||
| “ | Tanzania | Same | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Moshi | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Makuiuny | 0 | 80 | 0 | 0 | 0 | 20 | ✓ | 100 | “ | |
| “ | Tanzania | Arusha | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Karatu | 0 | 80 | 0 | 0 | 0 | 20 | ✓ | 85 | “ | |
| “ | Tanzania combined | 62 | “ | |||||||||
| “ | Tanzania | ✓ | (Scott et al. 2014a) | |||||||||
| Australia | Australia | Ipswich | 0 | 44 | 70 | 2 | 0 | 7 | 92 | 70 | ✓ | (Hamm and Scott 2009) |
| “ | “ | Bowhill | 0 | ✓ | ✓ | 0 | ✓ | ✓ | (Wagoner 1969b) | |||
| Asia | Japan | Furano | 0 | 0 | 9 | 0 | 0 | 91 | 0 | 0 | (Tomita and Wada 1989b) | |
| “ | “ | Sapporo | 0 | 0 | 29 | 0 | 0.6 | 70 | 0.3 | 0.6 | “ | |
| “ | “ | Akkeshi | 0 | 0 | 21 | 0 | 0 | 79 | 0 | 0 | “ | |
| “ | “ | Obihiro | 0 | 0 | 12 | 0 | 0 | 88 | 0 | 0 | “ | |
| “ | “ | Hachinohe | 0 | 0 | 38 | 0 | 0.5 | 57 | 2 | 4 | “ | |
| “ | “ | Niharu | 5 | 0 | 32 | 0 | 0 | 64 | 0 | “ | ||
| “ | “ | Togakushi | 0 | 8 | 58 | 0 | 0 | 35 | 4 | 28 | “ | |
| “ | “ | Haga | 0 | 2 | 96 | 0 | 0 | 2 | 4 | 17 | “ | |
| “ | “ | Miyagi | 0 | 3 | 35 | 0 | 0 | 63 | 5 | 0 | “ | |
| “ | “ | Hokota | 0 | 2 | 57 | 0 | 0 | 40 | 5 | “ | ||
| “ | “ | Kofu | 0 | 0 | 70 | 0 | 0 | 30 | 24 | 29 | “ | |
| “ | “ | Yumenoshima | 0 | 1 | 74 | 0 | 0 | 25 | 68 | 48 | 99 | “ |
| “ | “ | Aio | 2 | 31 | 29 | 3 | 22 | 12 | 2 | 1 | “ | |
| “ | “ | Kasuya | 1 | 16 | 39 | 0 | 18 | 26 | 20 | 38 | “ | |
| “ | “ | Nangoku | 3 | 0 | 24 | 0 | 3 | 70 | 6 | 13 | “ | |
| “ | “ | Haruno | 0 | 0 | 34 | 0 | 0 | 66 | 2 | 0 | “ | |
| “ | “ | Hachijo | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | Okinawa | 4 | 41 | 48 | 2 | 0 | 4 | 15 | 47 | “ | |
| “ | “ | Ishigaki | 0 | 32 | 54 | 0 | 0 | 14 | 2 | 4 | “ | |
| “ | “ | Kirishima | 0 | (Hiroyoshi 1964) | ||||||||
| “ | “ | Nichinan | 0 | “ | ||||||||
| “ | “ | Sakurai | 0 | “ | ||||||||
| “ | “ | Kitakyushu | ✓ | ✓ | ✓ | 0 | 0 | ✓* | ✓ | 0 | (Tsukamoto et al. 1980) | |
| “ | “ | Kitakyushu | 0 | ✓ | ✓ | 0 | ✓ | ✓* | ✓ | 0 | “ | |
| “ | “ | FR 83 | 80 | 0 | 0 | (Tomita and Wada 1989a) | ||||||
| “ | “ | OH 83 | 0 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | AK 83 | 0 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-YG 83 | 0 | 0 | 40 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-YG 84 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-OD 84 | 0 | 0 | 33 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | IK-RS 84 | 0 | 0 | 24 | 0 | 0 | 0 | 2 | 3 | “ | |
| “ | “ | IK-YU 84 | 0 | 0 | 30 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | IK-BN 84 | 0 | 0 | 35 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | OT-ZB 84 | 0 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | “ | |
| Osaka | ✓ | “ | ||||||||||
| Asia/Europe | Turkey | Giresun | ✓ | ✓ | (Cakir 1999) | |||||||
| “ | “ | Ordu | ✓ | ✓ | “ | |||||||
| “ | “ | Trabzon | ✓ | ✓ | “ | |||||||
| “ | “ | Giresun | 0 | ✓ | 0 | ✓ | (Cakir and Kence 2000) | |||||
| “ | “ | Trabzon | 0 | 0 | ✓ | ✓ | “ | |||||
| “ | “ | Kayrak | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Simav | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Izmit | ✓ | ✓ | 0 | ✓ | “ | |||||
| “ | “ | Iskenderun | ✓ | ✓ | ✓ | ✓ | “ | |||||
| “ | “ | Balikesirr | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Polatli | 0 | 0 | 0 | “ | ||||||
| Trabzon | ✓ | (Scott et al. 2014a) | ||||||||||
| Europe | British Isles | Fm31 | 0 | 0 | 0 | 0 | 0* | ✓ | ✓ | (Denholm et al. 1985) | ||
| “ | “ | Fm39 | 0 | 0 | ✓ | 0 | ✓ * | ✓ | “ | |||
| “ | “ | Fm42 | 0 | 0 | 0 | 0 | ✓ | “ | ||||
| “ | “ | Harpenden | ✓ | ✓ | ||||||||
| “ | “ | Fm44 | 0 | 0 | ✓ | 0 | ✓ | 25 | 35-52 | “ | ||
| “ | “ | Fm45 | 0 | 0 | ✓ | 0 | ✓ | ✓ | “ | |||
| “ | England | Fm6 | 0 | 0 | 6 | 0 | 0 | 94* | 6 | ✓ | (Denholm et al. 1983) | |
| “ | “ | Fm22 | 0 | 0 | 2.9 | 0 | 0 | 69* | 29 | ✓ | “ | |
| “ | Italy | 12 populations | 0 | 0 | 0 | 0 | 0 | ✓ | (Franco et al. 1982) | |||
| “ | “ | 11 populations | ✓ | ✓ | ✓ | ✓ | “ | |||||
| “ | “ | IT1 | 0 | 0 | 12 | 0 | 0 | 52 | 44 | (Kozielska et al. 2008) | ||
| “ | “ | IT2 | 0 | 25 | 9 | 0 | 0 | 44 | 43 | “ | ||
| “ | “ | IT3 | 0 | 0 | 0 | 0 | 0 | 50 | 10 | “ | ||
| “ | “ | IT4 | 12 | 9 | 45 | 0 | 0 | 42 | ✓ | 100 | “ | |
| “ | “ | IT5 | 2 | 17 | 50 | 0 | 9 | 62 | ✓ | 100 | “ | |
| “ | “ | IT6 | 3 | 13 | 32 | 0 | 0 | 68 | ✓ | 95 | “ | |
| “ | “ | IT7 | 0 | 3 | 53 | 0 | 0 | 17 | 78 | “ | ||
| “ | “ | IT8 | 9 | 3 | 86 | 3 | 3 | 16 | ✓ | 100 | “ | |
| “ | “ | IT9 | 8 | 17 | 46 | 0 | 0 | 6 | 86 | “ | ||
| “ | “ | IT10 | 3 | 0 | 55 | 3 | 0 | 0 | 95 | “ | ||
| “ | “ | IT11 | 0 | 0 | 76 | 0 | 0 | 3 | 96 | “ | ||
| “ | “ | IT12 | 0 | 0 | 56 | 0 | 0 | 8 | 47 | “ | ||
| “ | Switzerland | Switzerland | 0 | 0 | 0 | 0 | 0 | 50 | 5 | “ | ||
| “ | Germany | GE1 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | “ | ||
| “ | “ | GE2 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | “ | ||
| France | Faverges | ✓ | (Scott et al. 2014a) | |||||||||
| Spain | Santa Fé | ✓ | “ | |||||||||
| N. America | USA | Texas | 10 | 0 | ✓ | (McDonald and Overland 1974) | ||||||
| “ | “ | North Dakota | 8 | 0 | ✓ | “ | ||||||
| “ | “ | Florida | 100 | 0 | 0 | “ | ||||||
| “ | “ | Florida | 0 | 0 | 100 | 0 | 0 | 0 | (Hamm et al. 2005) | |||
| “ | “ | North Carolina 2002 | 0 | 0 | 20 | 0 | 0 | 78 | 2.4 | 0 | “ | |
| “ | “ | North Carolina 2006 | 0 | 0 | 19 | 0 | 0 | 78 | 1.4 | 1.4 | (Hamm and Scott 2008) | |
| “ | “ | North Carolina 2007 | 0 | 0 | 2.3 | 0 | 0 | 95 | 0 | 2.3 | 4.2 | “ |
| “ | “ | New York | 0 | 0 | 4.4 | 0 | 0 | 96 | (Hamm et al. 2005) | |||
| “ | “ | Maine | 0 | 0 | 0 | 0 | 0 | 100 | “ | |||
| “ | “ | California- Chino | 0 | 0 | 15 | 0 | 0 | 85 | ✓ | ✓ | (T. Shono and J. G. Scott, personal communication) | |
Blank cells indicate no information available (i.e., experiments not conducted or marker strain for specific autosome not used). ✓, detected, but not quantified.
Values can vary from one study to another primarily based on how males with multiple M factors were categorized. See the individual papers for details.
YM values from some studies indicate only that M was not linked to an autosome, thus linkage of M to Y or to X are possible in some of these studies.
Percentage of males being heterozygous for M at more than one linkage group (e.g., IIM/II; IIIM/III or IIIM/III; XYM). Zeroes indicate that appropriate methods for detection were used and that none were found.
Percentage of males producing only male offspring (i.e., homozygous for at least one autosome (AM/AM, XM/YM, or XM/XM). Zeroes indicate that appropriate methods for detection were used and that none were found.
Populations that have homozygous M males can be reasonably assumed to have Md-traD females. However, these cells were left blank unless there was detection (✓) or quantification of Md-traD.
XM males were found most commonly in this population (male determining factor did not map to an autosome and a male had a karyotype of XX).
| Location . | IM . | IIM . | IIIM . | IVM . | VM . | YMb . | M/+c . | M/Md . | Md-traDe . | Reference . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Africa | S. Africa | Johannesburg-Pretoria area SA1 | 0 | 0 | 100 | 0 | 0 | ✓ | ✓ | (Denholm et al. 1990) | ||
| “ | S. Africa | Johannesburg-Pretoria area SA2 | 0 | 0 | 85 | 0 | 7.4 | 7.4 | 45 | ✓ | “ | |
| “ | S. Africa | Zinkwazi Beach | 0 | ✓ | ✓ | 0 | 0 | 0 | 29 | (Feldmeyer et al. 2008) | ||
| “ | S. Africa | Umhlali | ✓ | ✓ | ✓ | 0 | ✓ | 0 | 79 | “ | ||
| “ | S. Africa | Hammarsdale | 0 | ✓ | ✓ | 0 | 0 | 0 | 92 | “ | ||
| “ | S. Africa | Ashburton | ✓ | ✓ | 0 | 0 | ✓ | 0 | 13 | “ | ||
| “ | S. Africa | Mooi River | 0 | ✓ | ✓ | 0 | 0 | 0 | 29 | “ | ||
| “ | S. Africa | Warden | 0 | 0 | 70 | 0 | 0 | 30 | ✓ | 15 | “ | |
| “ | S. Africa | South Africa combined | 26 | “ | ||||||||
| “ | Tanzania | Same | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Moshi | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Makuiuny | 0 | 80 | 0 | 0 | 0 | 20 | ✓ | 100 | “ | |
| “ | Tanzania | Arusha | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Karatu | 0 | 80 | 0 | 0 | 0 | 20 | ✓ | 85 | “ | |
| “ | Tanzania combined | 62 | “ | |||||||||
| “ | Tanzania | ✓ | (Scott et al. 2014a) | |||||||||
| Australia | Australia | Ipswich | 0 | 44 | 70 | 2 | 0 | 7 | 92 | 70 | ✓ | (Hamm and Scott 2009) |
| “ | “ | Bowhill | 0 | ✓ | ✓ | 0 | ✓ | ✓ | (Wagoner 1969b) | |||
| Asia | Japan | Furano | 0 | 0 | 9 | 0 | 0 | 91 | 0 | 0 | (Tomita and Wada 1989b) | |
| “ | “ | Sapporo | 0 | 0 | 29 | 0 | 0.6 | 70 | 0.3 | 0.6 | “ | |
| “ | “ | Akkeshi | 0 | 0 | 21 | 0 | 0 | 79 | 0 | 0 | “ | |
| “ | “ | Obihiro | 0 | 0 | 12 | 0 | 0 | 88 | 0 | 0 | “ | |
| “ | “ | Hachinohe | 0 | 0 | 38 | 0 | 0.5 | 57 | 2 | 4 | “ | |
| “ | “ | Niharu | 5 | 0 | 32 | 0 | 0 | 64 | 0 | “ | ||
| “ | “ | Togakushi | 0 | 8 | 58 | 0 | 0 | 35 | 4 | 28 | “ | |
| “ | “ | Haga | 0 | 2 | 96 | 0 | 0 | 2 | 4 | 17 | “ | |
| “ | “ | Miyagi | 0 | 3 | 35 | 0 | 0 | 63 | 5 | 0 | “ | |
| “ | “ | Hokota | 0 | 2 | 57 | 0 | 0 | 40 | 5 | “ | ||
| “ | “ | Kofu | 0 | 0 | 70 | 0 | 0 | 30 | 24 | 29 | “ | |
| “ | “ | Yumenoshima | 0 | 1 | 74 | 0 | 0 | 25 | 68 | 48 | 99 | “ |
| “ | “ | Aio | 2 | 31 | 29 | 3 | 22 | 12 | 2 | 1 | “ | |
| “ | “ | Kasuya | 1 | 16 | 39 | 0 | 18 | 26 | 20 | 38 | “ | |
| “ | “ | Nangoku | 3 | 0 | 24 | 0 | 3 | 70 | 6 | 13 | “ | |
| “ | “ | Haruno | 0 | 0 | 34 | 0 | 0 | 66 | 2 | 0 | “ | |
| “ | “ | Hachijo | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | Okinawa | 4 | 41 | 48 | 2 | 0 | 4 | 15 | 47 | “ | |
| “ | “ | Ishigaki | 0 | 32 | 54 | 0 | 0 | 14 | 2 | 4 | “ | |
| “ | “ | Kirishima | 0 | (Hiroyoshi 1964) | ||||||||
| “ | “ | Nichinan | 0 | “ | ||||||||
| “ | “ | Sakurai | 0 | “ | ||||||||
| “ | “ | Kitakyushu | ✓ | ✓ | ✓ | 0 | 0 | ✓* | ✓ | 0 | (Tsukamoto et al. 1980) | |
| “ | “ | Kitakyushu | 0 | ✓ | ✓ | 0 | ✓ | ✓* | ✓ | 0 | “ | |
| “ | “ | FR 83 | 80 | 0 | 0 | (Tomita and Wada 1989a) | ||||||
| “ | “ | OH 83 | 0 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | AK 83 | 0 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-YG 83 | 0 | 0 | 40 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-YG 84 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-OD 84 | 0 | 0 | 33 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | IK-RS 84 | 0 | 0 | 24 | 0 | 0 | 0 | 2 | 3 | “ | |
| “ | “ | IK-YU 84 | 0 | 0 | 30 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | IK-BN 84 | 0 | 0 | 35 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | OT-ZB 84 | 0 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | “ | |
| Osaka | ✓ | “ | ||||||||||
| Asia/Europe | Turkey | Giresun | ✓ | ✓ | (Cakir 1999) | |||||||
| “ | “ | Ordu | ✓ | ✓ | “ | |||||||
| “ | “ | Trabzon | ✓ | ✓ | “ | |||||||
| “ | “ | Giresun | 0 | ✓ | 0 | ✓ | (Cakir and Kence 2000) | |||||
| “ | “ | Trabzon | 0 | 0 | ✓ | ✓ | “ | |||||
| “ | “ | Kayrak | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Simav | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Izmit | ✓ | ✓ | 0 | ✓ | “ | |||||
| “ | “ | Iskenderun | ✓ | ✓ | ✓ | ✓ | “ | |||||
| “ | “ | Balikesirr | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Polatli | 0 | 0 | 0 | “ | ||||||
| Trabzon | ✓ | (Scott et al. 2014a) | ||||||||||
| Europe | British Isles | Fm31 | 0 | 0 | 0 | 0 | 0* | ✓ | ✓ | (Denholm et al. 1985) | ||
| “ | “ | Fm39 | 0 | 0 | ✓ | 0 | ✓ * | ✓ | “ | |||
| “ | “ | Fm42 | 0 | 0 | 0 | 0 | ✓ | “ | ||||
| “ | “ | Harpenden | ✓ | ✓ | ||||||||
| “ | “ | Fm44 | 0 | 0 | ✓ | 0 | ✓ | 25 | 35-52 | “ | ||
| “ | “ | Fm45 | 0 | 0 | ✓ | 0 | ✓ | ✓ | “ | |||
| “ | England | Fm6 | 0 | 0 | 6 | 0 | 0 | 94* | 6 | ✓ | (Denholm et al. 1983) | |
| “ | “ | Fm22 | 0 | 0 | 2.9 | 0 | 0 | 69* | 29 | ✓ | “ | |
| “ | Italy | 12 populations | 0 | 0 | 0 | 0 | 0 | ✓ | (Franco et al. 1982) | |||
| “ | “ | 11 populations | ✓ | ✓ | ✓ | ✓ | “ | |||||
| “ | “ | IT1 | 0 | 0 | 12 | 0 | 0 | 52 | 44 | (Kozielska et al. 2008) | ||
| “ | “ | IT2 | 0 | 25 | 9 | 0 | 0 | 44 | 43 | “ | ||
| “ | “ | IT3 | 0 | 0 | 0 | 0 | 0 | 50 | 10 | “ | ||
| “ | “ | IT4 | 12 | 9 | 45 | 0 | 0 | 42 | ✓ | 100 | “ | |
| “ | “ | IT5 | 2 | 17 | 50 | 0 | 9 | 62 | ✓ | 100 | “ | |
| “ | “ | IT6 | 3 | 13 | 32 | 0 | 0 | 68 | ✓ | 95 | “ | |
| “ | “ | IT7 | 0 | 3 | 53 | 0 | 0 | 17 | 78 | “ | ||
| “ | “ | IT8 | 9 | 3 | 86 | 3 | 3 | 16 | ✓ | 100 | “ | |
| “ | “ | IT9 | 8 | 17 | 46 | 0 | 0 | 6 | 86 | “ | ||
| “ | “ | IT10 | 3 | 0 | 55 | 3 | 0 | 0 | 95 | “ | ||
| “ | “ | IT11 | 0 | 0 | 76 | 0 | 0 | 3 | 96 | “ | ||
| “ | “ | IT12 | 0 | 0 | 56 | 0 | 0 | 8 | 47 | “ | ||
| “ | Switzerland | Switzerland | 0 | 0 | 0 | 0 | 0 | 50 | 5 | “ | ||
| “ | Germany | GE1 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | “ | ||
| “ | “ | GE2 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | “ | ||
| France | Faverges | ✓ | (Scott et al. 2014a) | |||||||||
| Spain | Santa Fé | ✓ | “ | |||||||||
| N. America | USA | Texas | 10 | 0 | ✓ | (McDonald and Overland 1974) | ||||||
| “ | “ | North Dakota | 8 | 0 | ✓ | “ | ||||||
| “ | “ | Florida | 100 | 0 | 0 | “ | ||||||
| “ | “ | Florida | 0 | 0 | 100 | 0 | 0 | 0 | (Hamm et al. 2005) | |||
| “ | “ | North Carolina 2002 | 0 | 0 | 20 | 0 | 0 | 78 | 2.4 | 0 | “ | |
| “ | “ | North Carolina 2006 | 0 | 0 | 19 | 0 | 0 | 78 | 1.4 | 1.4 | (Hamm and Scott 2008) | |
| “ | “ | North Carolina 2007 | 0 | 0 | 2.3 | 0 | 0 | 95 | 0 | 2.3 | 4.2 | “ |
| “ | “ | New York | 0 | 0 | 4.4 | 0 | 0 | 96 | (Hamm et al. 2005) | |||
| “ | “ | Maine | 0 | 0 | 0 | 0 | 0 | 100 | “ | |||
| “ | “ | California- Chino | 0 | 0 | 15 | 0 | 0 | 85 | ✓ | ✓ | (T. Shono and J. G. Scott, personal communication) | |
| Location . | IM . | IIM . | IIIM . | IVM . | VM . | YMb . | M/+c . | M/Md . | Md-traDe . | Reference . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Africa | S. Africa | Johannesburg-Pretoria area SA1 | 0 | 0 | 100 | 0 | 0 | ✓ | ✓ | (Denholm et al. 1990) | ||
| “ | S. Africa | Johannesburg-Pretoria area SA2 | 0 | 0 | 85 | 0 | 7.4 | 7.4 | 45 | ✓ | “ | |
| “ | S. Africa | Zinkwazi Beach | 0 | ✓ | ✓ | 0 | 0 | 0 | 29 | (Feldmeyer et al. 2008) | ||
| “ | S. Africa | Umhlali | ✓ | ✓ | ✓ | 0 | ✓ | 0 | 79 | “ | ||
| “ | S. Africa | Hammarsdale | 0 | ✓ | ✓ | 0 | 0 | 0 | 92 | “ | ||
| “ | S. Africa | Ashburton | ✓ | ✓ | 0 | 0 | ✓ | 0 | 13 | “ | ||
| “ | S. Africa | Mooi River | 0 | ✓ | ✓ | 0 | 0 | 0 | 29 | “ | ||
| “ | S. Africa | Warden | 0 | 0 | 70 | 0 | 0 | 30 | ✓ | 15 | “ | |
| “ | S. Africa | South Africa combined | 26 | “ | ||||||||
| “ | Tanzania | Same | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Moshi | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Makuiuny | 0 | 80 | 0 | 0 | 0 | 20 | ✓ | 100 | “ | |
| “ | Tanzania | Arusha | 0 | 100 | 0 | 0 | 0 | 0 | 100 | “ | ||
| “ | Tanzania | Karatu | 0 | 80 | 0 | 0 | 0 | 20 | ✓ | 85 | “ | |
| “ | Tanzania combined | 62 | “ | |||||||||
| “ | Tanzania | ✓ | (Scott et al. 2014a) | |||||||||
| Australia | Australia | Ipswich | 0 | 44 | 70 | 2 | 0 | 7 | 92 | 70 | ✓ | (Hamm and Scott 2009) |
| “ | “ | Bowhill | 0 | ✓ | ✓ | 0 | ✓ | ✓ | (Wagoner 1969b) | |||
| Asia | Japan | Furano | 0 | 0 | 9 | 0 | 0 | 91 | 0 | 0 | (Tomita and Wada 1989b) | |
| “ | “ | Sapporo | 0 | 0 | 29 | 0 | 0.6 | 70 | 0.3 | 0.6 | “ | |
| “ | “ | Akkeshi | 0 | 0 | 21 | 0 | 0 | 79 | 0 | 0 | “ | |
| “ | “ | Obihiro | 0 | 0 | 12 | 0 | 0 | 88 | 0 | 0 | “ | |
| “ | “ | Hachinohe | 0 | 0 | 38 | 0 | 0.5 | 57 | 2 | 4 | “ | |
| “ | “ | Niharu | 5 | 0 | 32 | 0 | 0 | 64 | 0 | “ | ||
| “ | “ | Togakushi | 0 | 8 | 58 | 0 | 0 | 35 | 4 | 28 | “ | |
| “ | “ | Haga | 0 | 2 | 96 | 0 | 0 | 2 | 4 | 17 | “ | |
| “ | “ | Miyagi | 0 | 3 | 35 | 0 | 0 | 63 | 5 | 0 | “ | |
| “ | “ | Hokota | 0 | 2 | 57 | 0 | 0 | 40 | 5 | “ | ||
| “ | “ | Kofu | 0 | 0 | 70 | 0 | 0 | 30 | 24 | 29 | “ | |
| “ | “ | Yumenoshima | 0 | 1 | 74 | 0 | 0 | 25 | 68 | 48 | 99 | “ |
| “ | “ | Aio | 2 | 31 | 29 | 3 | 22 | 12 | 2 | 1 | “ | |
| “ | “ | Kasuya | 1 | 16 | 39 | 0 | 18 | 26 | 20 | 38 | “ | |
| “ | “ | Nangoku | 3 | 0 | 24 | 0 | 3 | 70 | 6 | 13 | “ | |
| “ | “ | Haruno | 0 | 0 | 34 | 0 | 0 | 66 | 2 | 0 | “ | |
| “ | “ | Hachijo | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | Okinawa | 4 | 41 | 48 | 2 | 0 | 4 | 15 | 47 | “ | |
| “ | “ | Ishigaki | 0 | 32 | 54 | 0 | 0 | 14 | 2 | 4 | “ | |
| “ | “ | Kirishima | 0 | (Hiroyoshi 1964) | ||||||||
| “ | “ | Nichinan | 0 | “ | ||||||||
| “ | “ | Sakurai | 0 | “ | ||||||||
| “ | “ | Kitakyushu | ✓ | ✓ | ✓ | 0 | 0 | ✓* | ✓ | 0 | (Tsukamoto et al. 1980) | |
| “ | “ | Kitakyushu | 0 | ✓ | ✓ | 0 | ✓ | ✓* | ✓ | 0 | “ | |
| “ | “ | FR 83 | 80 | 0 | 0 | (Tomita and Wada 1989a) | ||||||
| “ | “ | OH 83 | 0 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | AK 83 | 0 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-YG 83 | 0 | 0 | 40 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-YG 84 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | SP-OD 84 | 0 | 0 | 33 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | IK-RS 84 | 0 | 0 | 24 | 0 | 0 | 0 | 2 | 3 | “ | |
| “ | “ | IK-YU 84 | 0 | 0 | 30 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | IK-BN 84 | 0 | 0 | 35 | 0 | 0 | 0 | 0 | 0 | “ | |
| “ | “ | OT-ZB 84 | 0 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | “ | |
| Osaka | ✓ | “ | ||||||||||
| Asia/Europe | Turkey | Giresun | ✓ | ✓ | (Cakir 1999) | |||||||
| “ | “ | Ordu | ✓ | ✓ | “ | |||||||
| “ | “ | Trabzon | ✓ | ✓ | “ | |||||||
| “ | “ | Giresun | 0 | ✓ | 0 | ✓ | (Cakir and Kence 2000) | |||||
| “ | “ | Trabzon | 0 | 0 | ✓ | ✓ | “ | |||||
| “ | “ | Kayrak | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Simav | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Izmit | ✓ | ✓ | 0 | ✓ | “ | |||||
| “ | “ | Iskenderun | ✓ | ✓ | ✓ | ✓ | “ | |||||
| “ | “ | Balikesirr | 0 | ✓ | 0 | “ | ||||||
| “ | “ | Polatli | 0 | 0 | 0 | “ | ||||||
| Trabzon | ✓ | (Scott et al. 2014a) | ||||||||||
| Europe | British Isles | Fm31 | 0 | 0 | 0 | 0 | 0* | ✓ | ✓ | (Denholm et al. 1985) | ||
| “ | “ | Fm39 | 0 | 0 | ✓ | 0 | ✓ * | ✓ | “ | |||
| “ | “ | Fm42 | 0 | 0 | 0 | 0 | ✓ | “ | ||||
| “ | “ | Harpenden | ✓ | ✓ | ||||||||
| “ | “ | Fm44 | 0 | 0 | ✓ | 0 | ✓ | 25 | 35-52 | “ | ||
| “ | “ | Fm45 | 0 | 0 | ✓ | 0 | ✓ | ✓ | “ | |||
| “ | England | Fm6 | 0 | 0 | 6 | 0 | 0 | 94* | 6 | ✓ | (Denholm et al. 1983) | |
| “ | “ | Fm22 | 0 | 0 | 2.9 | 0 | 0 | 69* | 29 | ✓ | “ | |
| “ | Italy | 12 populations | 0 | 0 | 0 | 0 | 0 | ✓ | (Franco et al. 1982) | |||
| “ | “ | 11 populations | ✓ | ✓ | ✓ | ✓ | “ | |||||
| “ | “ | IT1 | 0 | 0 | 12 | 0 | 0 | 52 | 44 | (Kozielska et al. 2008) | ||
| “ | “ | IT2 | 0 | 25 | 9 | 0 | 0 | 44 | 43 | “ | ||
| “ | “ | IT3 | 0 | 0 | 0 | 0 | 0 | 50 | 10 | “ | ||
| “ | “ | IT4 | 12 | 9 | 45 | 0 | 0 | 42 | ✓ | 100 | “ | |
| “ | “ | IT5 | 2 | 17 | 50 | 0 | 9 | 62 | ✓ | 100 | “ | |
| “ | “ | IT6 | 3 | 13 | 32 | 0 | 0 | 68 | ✓ | 95 | “ | |
| “ | “ | IT7 | 0 | 3 | 53 | 0 | 0 | 17 | 78 | “ | ||
| “ | “ | IT8 | 9 | 3 | 86 | 3 | 3 | 16 | ✓ | 100 | “ | |
| “ | “ | IT9 | 8 | 17 | 46 | 0 | 0 | 6 | 86 | “ | ||
| “ | “ | IT10 | 3 | 0 | 55 | 3 | 0 | 0 | 95 | “ | ||
| “ | “ | IT11 | 0 | 0 | 76 | 0 | 0 | 3 | 96 | “ | ||
| “ | “ | IT12 | 0 | 0 | 56 | 0 | 0 | 8 | 47 | “ | ||
| “ | Switzerland | Switzerland | 0 | 0 | 0 | 0 | 0 | 50 | 5 | “ | ||
| “ | Germany | GE1 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | “ | ||
| “ | “ | GE2 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | “ | ||
| France | Faverges | ✓ | (Scott et al. 2014a) | |||||||||
| Spain | Santa Fé | ✓ | “ | |||||||||
| N. America | USA | Texas | 10 | 0 | ✓ | (McDonald and Overland 1974) | ||||||
| “ | “ | North Dakota | 8 | 0 | ✓ | “ | ||||||
| “ | “ | Florida | 100 | 0 | 0 | “ | ||||||
| “ | “ | Florida | 0 | 0 | 100 | 0 | 0 | 0 | (Hamm et al. 2005) | |||
| “ | “ | North Carolina 2002 | 0 | 0 | 20 | 0 | 0 | 78 | 2.4 | 0 | “ | |
| “ | “ | North Carolina 2006 | 0 | 0 | 19 | 0 | 0 | 78 | 1.4 | 1.4 | (Hamm and Scott 2008) | |
| “ | “ | North Carolina 2007 | 0 | 0 | 2.3 | 0 | 0 | 95 | 0 | 2.3 | 4.2 | “ |
| “ | “ | New York | 0 | 0 | 4.4 | 0 | 0 | 96 | (Hamm et al. 2005) | |||
| “ | “ | Maine | 0 | 0 | 0 | 0 | 0 | 100 | “ | |||
| “ | “ | California- Chino | 0 | 0 | 15 | 0 | 0 | 85 | ✓ | ✓ | (T. Shono and J. G. Scott, personal communication) | |
Blank cells indicate no information available (i.e., experiments not conducted or marker strain for specific autosome not used). ✓, detected, but not quantified.
Values can vary from one study to another primarily based on how males with multiple M factors were categorized. See the individual papers for details.
YM values from some studies indicate only that M was not linked to an autosome, thus linkage of M to Y or to X are possible in some of these studies.
Percentage of males being heterozygous for M at more than one linkage group (e.g., IIM/II; IIIM/III or IIIM/III; XYM). Zeroes indicate that appropriate methods for detection were used and that none were found.
Percentage of males producing only male offspring (i.e., homozygous for at least one autosome (AM/AM, XM/YM, or XM/XM). Zeroes indicate that appropriate methods for detection were used and that none were found.
Populations that have homozygous M males can be reasonably assumed to have Md-traD females. However, these cells were left blank unless there was detection (✓) or quantification of Md-traD.
XM males were found most commonly in this population (male determining factor did not map to an autosome and a male had a karyotype of XX).
It was shown recently that the gray flesh fly, Sarcophaga bullata, X chromosome is homologous to the Drosophila “dot” chromosome (chromosome 4 in D. melanogaster), and this chromosome is likely to be the ancestral X chromosome of Brachycera (Vicoso and Bachtrog 2013). The house fly sex chromosomes, therefore, likely reflect an ancient X/Y pair, and decreases in chromosome number among the Muscidae are likely the result of fusions of the ancestral sex chromosomes with one of the five autosomes.
“Autosomal” (AM) or “atypical” (Rubini et al. 1980) house fly strains have the M factor located on one or more of the five autosomes (I−V) or the X (Note: The AM designation is a bit misleading because XM males also are considered “atypical”). The M factor has been shown to have varying degrees of strength depending on its location (Schmidt et al. 1997b). IM males are weak intersexes expressing female-specific yolk proteins (Schmidt et al. 1997b) and both the male and female isoforms of Md-dsx (Siegenthaler et al. 2009). The suggested cause of this is the presence of prominent stretches of heterochromatin on autosome I (Hediger et al. 1998; Dübendorfer 2001). YM (if multiple copies of M are present), IIIM, and VM show strong male-determining effects in the soma and impede the activity of Md-tra when introduced into the female germline by transplantation of progenitor germline cells (Schmidt et al. 1997b).
It is hypothesized that AM or XM factors are the result of transposition of the Y-linked M (Hiroyoshi 1964), and several lines of evidence suggest that M “inserts” into a single location on each autosome or X (Inoue and Hiroyoshi 1986). House flies in South East England contained a high frequency of XM individuals and “a small secondary constriction on X appeared to indicate reliably the presence of XM” (Denholm et al. 1983). The linkage of M was investigated using three IM strains and two IIIM strains collected in Japan (Inoue et al. 1983). All three IM factors mapped to the right of the black puparium (bp) gene, and M was found tightly linked to pointed wings (pw) in both IIIM strains, suggesting that M occupies a definite site on the respective chromosomes (Inoue et al. 1983). The authors concluded that AM factors are located in centric heterochromatin on each autosome and M factors on a given chromosome are all at the same locus. Alternatively, it was proposed that the M factors on the different autosomes are different genes that adopt the function of male-determiner through mutation (Bopp 2010).
Md-tra is located on autosome IV and has two different functional variants. The “wild-type” allele is sensitive to inhibition by M, whereas a dominant allele (Md-traD, formerly FD) is resistant to M and acts as a female-determining factor (McDonald et al. 1978; Inoue and Hiroyoshi 1986; Cakir 1999; Hediger et al. 2010) (Figure 1E). The Md-traD allele may have invaded natural populations because of sex ratio selection (Hamilton 1967; Bull and Charnov 1977; Kozielska et al. 2006). Populations that contain males with multiple M factors (IIIM and YM, for example) or males homozygous for an AM can skew the sex ratio away from 1:1 male:female. The presence of Md-traD in the zygotic genotype causes female development even in the presence of up to three M factors (McDonald et al. 1978; Schmidt et al. 1997b; Hediger et al. 1998), potentially balancing the sex ratio in populations with multiple M factors. In populations in which males are exclusively AM/AM and the Md-traD allele segregates (Franco et al. 1982; Denholm et al. 1983, 1985; Denholm et al. 1990), females are the heterogametic sex (Md-traD/Md-tra+). Md-traD has been reported in populations from Africa, Asia, Australia, Europe, and North America (Table 1).
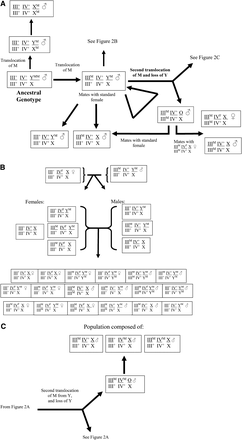
Figure 2 presents a hypothetical general scheme of the changes that are likely to occur as a house fly population evolves from one type of sex determination system to one of the others. The scheme represented in Figure 2 assumes that M can be mobilized from Y to another chromosome (once or twice) with the resulting loss (over time) of the Y chromosome. The proposed scheme accounts for genotypes found in nature, although some karyotypes that rarely have been detected (Table 2) are not included for the sake of simplicity.
Schematic representation of the evolution of changes in the linkage of M and frequency of F (Md-traD) in the house fly, M. domestica. Autosomes III and IV are used for illustration purposes but could be any of the autosomes (see Table 1). Genotype is given only for the male unless otherwise specified. Females are assumed to be III+/III+ ; IV+/IV+; XX unless otherwise specified. (A) Changes possible from the ancestral state (XYM, with no autosomomal males). (B) Continued from (A). Schematic representation of the evolution of males and females homozygous for M. (C) Continued from (A). Schematic representation of the evolution of males with copies of M on different autosomes. The AM factors are assumed to be derived from the M factor on Y (Hiroyoshi 1964), and the M factors are thought to incorporate into a specific site on each autosome (Inoue et al. 1983).
Percentage of male house flies with specific karyotypes
| . | . | . | . | Percentage . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Location . | n . | XY . | XX . | XO . | OY . | XXX . | XXY . | YY . | Reference . | ||
| Africa | S. Africa | SA1 | 31 | 90 | 10 | (Denholm et al. 1990) | |||||
| “ | “ | SA2 | 33 | 30 | 64 | 6 | “ | ||||
| Asia/Europe | Turkey | Antalya | 30 | 53 | 47 | (Cakir and Kence 1996) | |||||
| “ | “ | Incekum | 31 | 74 | 26 | “ | |||||
| “ | “ | Anamur | 31 | 77 | 23 | “ | |||||
| “ | “ | Gulnar | 32 | 34 | 66 | “ | |||||
| “ | “ | Kayrak | 30 | 10 | 90 | “ | |||||
| “ | “ | Y. Cadiri | 32 | 41 | 59 | “ | |||||
| “ | “ | Silifke | 30 | 30 | 70 | “ | |||||
| “ | “ | Atakent | 30 | 60 | 40 | “ | |||||
| “ | “ | Mersin | 36 | 42 | 58 | “ | |||||
| “ | “ | Adana | 30 | 43 | 57 | “ | |||||
| “ | “ | Yumurtalik | 30 | 23 | 77 | “ | |||||
| “ | “ | Karatas | 25 | 56 | 44 | “ | |||||
| “ | “ | Ceyhan | 30 | 70 | 30 | “ | |||||
| “ | “ | Samsun | 32 | 75 | 25 | “ | |||||
| “ | “ | Giresun | 30 | 0 | 100 | “ | |||||
| “ | “ | Trabzon | 33 | 0 | 100 | “ | |||||
| “ | “ | Rize | 30 | 13 | 87 | “ | |||||
| “ | “ | Artvin | 33 | 24 | 76 | “ | |||||
| “ | “ | Erzurum | 32 | 53 | 47 | “ | |||||
| “ | “ | Erzincan | 36 | 83 | 17 | “ | |||||
| “ | “ | Sivas | 33 | 91 | 9 | “ | |||||
| “ | “ | Yozgat | 30 | 77 | 23 | “ | |||||
| “ | “ | Izmit | 30 | 100 | “ | ||||||
| “ | “ | Isparta | 30 | 40 | 60 | “ | |||||
| “ | “ | Bursa | 31 | 68 | 32 | “ | |||||
| “ | “ | Tokat | 36 | 69 | 31 | “ | |||||
| “ | “ | Istanbul | 30 | 23 | 77 | “ | |||||
| “ | “ | Iskenderun | 29 | 24 | 76 | “ | |||||
| “ | “ | Afyon | 28 | 61 | 39 | “ | |||||
| “ | “ | Usak | 32 | 50 | 50 | “ | |||||
| “ | “ | Ismir | 31 | 29 | 71 | “ | |||||
| “ | “ | Manisa | 29 | 55 | 45 | “ | |||||
| “ | “ | Balikesir | 28 | 25 | 75 | “ | |||||
| “ | “ | Simav | 29 | 30 | 70 | “ | |||||
| “ | “ | Ankara | 31 | 71 | 29 | “ | |||||
| “ | “ | Polatli | 31 | 97 | 3 | “ | |||||
| Europe | UK | Fm 42 | 51 | 88 | 4 | 8 | (Denholm et al. 1985) | ||||
| “ | “ | Fm 39 | 33 | 58 | 39 | 3 | “ | ||||
| “ | “ | Fm 31 | 28 | 21 | 79 | “ | |||||
| “ | “ | Fm 44 | 47 | 98 | 2 | “ | |||||
| “ | “ | Fm 45 | 48 | 75 | 19 | 4 | 2 | “ | |||
| “ | “ | Harpenden | 223 | 5 | 93 | 2 | 0 | “ | |||
| “ | “ | Fm 3 | 19 | 21 | 79 | (Denholm et al. 1983) | |||||
| “ | “ | Fm 9 | 33 | 100 | “ | ||||||
| “ | “ | Fm 6 | 36 | 89 | 11 | “ | |||||
| “ | “ | Fm 13 | 29 | 100 | “ | ||||||
| “ | “ | Fm 11 | 11 | 100 | “ | ||||||
| “ | “ | Fm 14 | 27 | 11 | 89 | “ | |||||
| “ | “ | Fm 22 | 46 | 2 | 98 | “ | |||||
| “ | “ | Fm 29 | 22 | 14 | 82 | 5 | “ | ||||
| “ | France | M1 | 87 | 77 | 15 | 8 | (Franco et al. 1982) | ||||
| “ | France | M2 | 49 | 59 | 41 | “ | |||||
| “ | Yugoslavia | M3 | 69 | 46 | 52 | 1 | “ | ||||
| “ | Italy | M4 | 92 | 85 | 15 | “ | |||||
| “ | Italy | M5 (2r) | 178 | 88 | 12 | “ | |||||
| “ | Italy | M5 (2r) | 94 | 67 | 17 | 16 | “ | ||||
| “ | Italy | M6 | 44 | 50 | 50 | “ | |||||
| “ | Italy | M7 (2r) | 149 | 64 | 36 | 1 | “ | ||||
| “ | Italy | M8 | 56 | 84 | 7 | 9 | “ | ||||
| “ | Italy | M9 (2) | 52 | 83 | 15 | 2 | “ | ||||
| “ | Italy | M10 | 72 | 31 | 67 | 3 | “ | ||||
| “ | Italy | M11 | 46 | 4 | 96 | “ | |||||
| “ | Italy | M12 | 61 | 39 | 57 | 3 | “ | ||||
| “ | Italy | M13 | 63 | 2 | 92 | 6 | “ | ||||
| “ | Italy | M14 | 72 | 19 | 81 | “ | |||||
| “ | Italy | M15 | 43 | 56 | 44 | “ | |||||
| “ | Italy | M16 | 54 | 35 | 61 | 4 | “ | ||||
| “ | Italy | M17 | 62 | 2 | 98 | “ | |||||
| “ | Italy | M18 | 25 | 4 | 96 | “ | |||||
| “ | Italy | M19 | 40 | 15 | 85 | “ | |||||
| “ | Sardinia | M20 | 96 | 10 | 90 | “ | |||||
| “ | Sardinia | M21 | 68 | 9 | 91 | “ | |||||
| “ | Iceland | S1 | 30 | 100 | “ | ||||||
| “ | Denmark | S2-3-42 | 105 | 100 | “ | ||||||
| “ | Netherlands | S5-6r-7 | 162 | 100 | “ | ||||||
| “ | Germany | S8 | 85 | 100 | |||||||
| “ | Switzerland | S9-10-11 | 167 | 100 | “ | ||||||
| “ | Italy | A1-2-3-4-5 | 130 | 100 | “ | ||||||
| “ | Italy | A6-7-8-9 | 253 | 100 | “ | ||||||
| “ | Sicily | A10-11 | 83 | 100 | “ | ||||||
| Total | 4416 | 45 | 53 | 0.27 | 0.09 | 0.07 | 0.36 | 1.8 | |||
| . | . | . | . | Percentage . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Location . | n . | XY . | XX . | XO . | OY . | XXX . | XXY . | YY . | Reference . | ||
| Africa | S. Africa | SA1 | 31 | 90 | 10 | (Denholm et al. 1990) | |||||
| “ | “ | SA2 | 33 | 30 | 64 | 6 | “ | ||||
| Asia/Europe | Turkey | Antalya | 30 | 53 | 47 | (Cakir and Kence 1996) | |||||
| “ | “ | Incekum | 31 | 74 | 26 | “ | |||||
| “ | “ | Anamur | 31 | 77 | 23 | “ | |||||
| “ | “ | Gulnar | 32 | 34 | 66 | “ | |||||
| “ | “ | Kayrak | 30 | 10 | 90 | “ | |||||
| “ | “ | Y. Cadiri | 32 | 41 | 59 | “ | |||||
| “ | “ | Silifke | 30 | 30 | 70 | “ | |||||
| “ | “ | Atakent | 30 | 60 | 40 | “ | |||||
| “ | “ | Mersin | 36 | 42 | 58 | “ | |||||
| “ | “ | Adana | 30 | 43 | 57 | “ | |||||
| “ | “ | Yumurtalik | 30 | 23 | 77 | “ | |||||
| “ | “ | Karatas | 25 | 56 | 44 | “ | |||||
| “ | “ | Ceyhan | 30 | 70 | 30 | “ | |||||
| “ | “ | Samsun | 32 | 75 | 25 | “ | |||||
| “ | “ | Giresun | 30 | 0 | 100 | “ | |||||
| “ | “ | Trabzon | 33 | 0 | 100 | “ | |||||
| “ | “ | Rize | 30 | 13 | 87 | “ | |||||
| “ | “ | Artvin | 33 | 24 | 76 | “ | |||||
| “ | “ | Erzurum | 32 | 53 | 47 | “ | |||||
| “ | “ | Erzincan | 36 | 83 | 17 | “ | |||||
| “ | “ | Sivas | 33 | 91 | 9 | “ | |||||
| “ | “ | Yozgat | 30 | 77 | 23 | “ | |||||
| “ | “ | Izmit | 30 | 100 | “ | ||||||
| “ | “ | Isparta | 30 | 40 | 60 | “ | |||||
| “ | “ | Bursa | 31 | 68 | 32 | “ | |||||
| “ | “ | Tokat | 36 | 69 | 31 | “ | |||||
| “ | “ | Istanbul | 30 | 23 | 77 | “ | |||||
| “ | “ | Iskenderun | 29 | 24 | 76 | “ | |||||
| “ | “ | Afyon | 28 | 61 | 39 | “ | |||||
| “ | “ | Usak | 32 | 50 | 50 | “ | |||||
| “ | “ | Ismir | 31 | 29 | 71 | “ | |||||
| “ | “ | Manisa | 29 | 55 | 45 | “ | |||||
| “ | “ | Balikesir | 28 | 25 | 75 | “ | |||||
| “ | “ | Simav | 29 | 30 | 70 | “ | |||||
| “ | “ | Ankara | 31 | 71 | 29 | “ | |||||
| “ | “ | Polatli | 31 | 97 | 3 | “ | |||||
| Europe | UK | Fm 42 | 51 | 88 | 4 | 8 | (Denholm et al. 1985) | ||||
| “ | “ | Fm 39 | 33 | 58 | 39 | 3 | “ | ||||
| “ | “ | Fm 31 | 28 | 21 | 79 | “ | |||||
| “ | “ | Fm 44 | 47 | 98 | 2 | “ | |||||
| “ | “ | Fm 45 | 48 | 75 | 19 | 4 | 2 | “ | |||
| “ | “ | Harpenden | 223 | 5 | 93 | 2 | 0 | “ | |||
| “ | “ | Fm 3 | 19 | 21 | 79 | (Denholm et al. 1983) | |||||
| “ | “ | Fm 9 | 33 | 100 | “ | ||||||
| “ | “ | Fm 6 | 36 | 89 | 11 | “ | |||||
| “ | “ | Fm 13 | 29 | 100 | “ | ||||||
| “ | “ | Fm 11 | 11 | 100 | “ | ||||||
| “ | “ | Fm 14 | 27 | 11 | 89 | “ | |||||
| “ | “ | Fm 22 | 46 | 2 | 98 | “ | |||||
| “ | “ | Fm 29 | 22 | 14 | 82 | 5 | “ | ||||
| “ | France | M1 | 87 | 77 | 15 | 8 | (Franco et al. 1982) | ||||
| “ | France | M2 | 49 | 59 | 41 | “ | |||||
| “ | Yugoslavia | M3 | 69 | 46 | 52 | 1 | “ | ||||
| “ | Italy | M4 | 92 | 85 | 15 | “ | |||||
| “ | Italy | M5 (2r) | 178 | 88 | 12 | “ | |||||
| “ | Italy | M5 (2r) | 94 | 67 | 17 | 16 | “ | ||||
| “ | Italy | M6 | 44 | 50 | 50 | “ | |||||
| “ | Italy | M7 (2r) | 149 | 64 | 36 | 1 | “ | ||||
| “ | Italy | M8 | 56 | 84 | 7 | 9 | “ | ||||
| “ | Italy | M9 (2) | 52 | 83 | 15 | 2 | “ | ||||
| “ | Italy | M10 | 72 | 31 | 67 | 3 | “ | ||||
| “ | Italy | M11 | 46 | 4 | 96 | “ | |||||
| “ | Italy | M12 | 61 | 39 | 57 | 3 | “ | ||||
| “ | Italy | M13 | 63 | 2 | 92 | 6 | “ | ||||
| “ | Italy | M14 | 72 | 19 | 81 | “ | |||||
| “ | Italy | M15 | 43 | 56 | 44 | “ | |||||
| “ | Italy | M16 | 54 | 35 | 61 | 4 | “ | ||||
| “ | Italy | M17 | 62 | 2 | 98 | “ | |||||
| “ | Italy | M18 | 25 | 4 | 96 | “ | |||||
| “ | Italy | M19 | 40 | 15 | 85 | “ | |||||
| “ | Sardinia | M20 | 96 | 10 | 90 | “ | |||||
| “ | Sardinia | M21 | 68 | 9 | 91 | “ | |||||
| “ | Iceland | S1 | 30 | 100 | “ | ||||||
| “ | Denmark | S2-3-42 | 105 | 100 | “ | ||||||
| “ | Netherlands | S5-6r-7 | 162 | 100 | “ | ||||||
| “ | Germany | S8 | 85 | 100 | |||||||
| “ | Switzerland | S9-10-11 | 167 | 100 | “ | ||||||
| “ | Italy | A1-2-3-4-5 | 130 | 100 | “ | ||||||
| “ | Italy | A6-7-8-9 | 253 | 100 | “ | ||||||
| “ | Sicily | A10-11 | 83 | 100 | “ | ||||||
| Total | 4416 | 45 | 53 | 0.27 | 0.09 | 0.07 | 0.36 | 1.8 | |||
Blank cells equal 0%.
| . | . | . | . | Percentage . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Location . | n . | XY . | XX . | XO . | OY . | XXX . | XXY . | YY . | Reference . | ||
| Africa | S. Africa | SA1 | 31 | 90 | 10 | (Denholm et al. 1990) | |||||
| “ | “ | SA2 | 33 | 30 | 64 | 6 | “ | ||||
| Asia/Europe | Turkey | Antalya | 30 | 53 | 47 | (Cakir and Kence 1996) | |||||
| “ | “ | Incekum | 31 | 74 | 26 | “ | |||||
| “ | “ | Anamur | 31 | 77 | 23 | “ | |||||
| “ | “ | Gulnar | 32 | 34 | 66 | “ | |||||
| “ | “ | Kayrak | 30 | 10 | 90 | “ | |||||
| “ | “ | Y. Cadiri | 32 | 41 | 59 | “ | |||||
| “ | “ | Silifke | 30 | 30 | 70 | “ | |||||
| “ | “ | Atakent | 30 | 60 | 40 | “ | |||||
| “ | “ | Mersin | 36 | 42 | 58 | “ | |||||
| “ | “ | Adana | 30 | 43 | 57 | “ | |||||
| “ | “ | Yumurtalik | 30 | 23 | 77 | “ | |||||
| “ | “ | Karatas | 25 | 56 | 44 | “ | |||||
| “ | “ | Ceyhan | 30 | 70 | 30 | “ | |||||
| “ | “ | Samsun | 32 | 75 | 25 | “ | |||||
| “ | “ | Giresun | 30 | 0 | 100 | “ | |||||
| “ | “ | Trabzon | 33 | 0 | 100 | “ | |||||
| “ | “ | Rize | 30 | 13 | 87 | “ | |||||
| “ | “ | Artvin | 33 | 24 | 76 | “ | |||||
| “ | “ | Erzurum | 32 | 53 | 47 | “ | |||||
| “ | “ | Erzincan | 36 | 83 | 17 | “ | |||||
| “ | “ | Sivas | 33 | 91 | 9 | “ | |||||
| “ | “ | Yozgat | 30 | 77 | 23 | “ | |||||
| “ | “ | Izmit | 30 | 100 | “ | ||||||
| “ | “ | Isparta | 30 | 40 | 60 | “ | |||||
| “ | “ | Bursa | 31 | 68 | 32 | “ | |||||
| “ | “ | Tokat | 36 | 69 | 31 | “ | |||||
| “ | “ | Istanbul | 30 | 23 | 77 | “ | |||||
| “ | “ | Iskenderun | 29 | 24 | 76 | “ | |||||
| “ | “ | Afyon | 28 | 61 | 39 | “ | |||||
| “ | “ | Usak | 32 | 50 | 50 | “ | |||||
| “ | “ | Ismir | 31 | 29 | 71 | “ | |||||
| “ | “ | Manisa | 29 | 55 | 45 | “ | |||||
| “ | “ | Balikesir | 28 | 25 | 75 | “ | |||||
| “ | “ | Simav | 29 | 30 | 70 | “ | |||||
| “ | “ | Ankara | 31 | 71 | 29 | “ | |||||
| “ | “ | Polatli | 31 | 97 | 3 | “ | |||||
| Europe | UK | Fm 42 | 51 | 88 | 4 | 8 | (Denholm et al. 1985) | ||||
| “ | “ | Fm 39 | 33 | 58 | 39 | 3 | “ | ||||
| “ | “ | Fm 31 | 28 | 21 | 79 | “ | |||||
| “ | “ | Fm 44 | 47 | 98 | 2 | “ | |||||
| “ | “ | Fm 45 | 48 | 75 | 19 | 4 | 2 | “ | |||
| “ | “ | Harpenden | 223 | 5 | 93 | 2 | 0 | “ | |||
| “ | “ | Fm 3 | 19 | 21 | 79 | (Denholm et al. 1983) | |||||
| “ | “ | Fm 9 | 33 | 100 | “ | ||||||
| “ | “ | Fm 6 | 36 | 89 | 11 | “ | |||||
| “ | “ | Fm 13 | 29 | 100 | “ | ||||||
| “ | “ | Fm 11 | 11 | 100 | “ | ||||||
| “ | “ | Fm 14 | 27 | 11 | 89 | “ | |||||
| “ | “ | Fm 22 | 46 | 2 | 98 | “ | |||||
| “ | “ | Fm 29 | 22 | 14 | 82 | 5 | “ | ||||
| “ | France | M1 | 87 | 77 | 15 | 8 | (Franco et al. 1982) | ||||
| “ | France | M2 | 49 | 59 | 41 | “ | |||||
| “ | Yugoslavia | M3 | 69 | 46 | 52 | 1 | “ | ||||
| “ | Italy | M4 | 92 | 85 | 15 | “ | |||||
| “ | Italy | M5 (2r) | 178 | 88 | 12 | “ | |||||
| “ | Italy | M5 (2r) | 94 | 67 | 17 | 16 | “ | ||||
| “ | Italy | M6 | 44 | 50 | 50 | “ | |||||
| “ | Italy | M7 (2r) | 149 | 64 | 36 | 1 | “ | ||||
| “ | Italy | M8 | 56 | 84 | 7 | 9 | “ | ||||
| “ | Italy | M9 (2) | 52 | 83 | 15 | 2 | “ | ||||
| “ | Italy | M10 | 72 | 31 | 67 | 3 | “ | ||||
| “ | Italy | M11 | 46 | 4 | 96 | “ | |||||
| “ | Italy | M12 | 61 | 39 | 57 | 3 | “ | ||||
| “ | Italy | M13 | 63 | 2 | 92 | 6 | “ | ||||
| “ | Italy | M14 | 72 | 19 | 81 | “ | |||||
| “ | Italy | M15 | 43 | 56 | 44 | “ | |||||
| “ | Italy | M16 | 54 | 35 | 61 | 4 | “ | ||||
| “ | Italy | M17 | 62 | 2 | 98 | “ | |||||
| “ | Italy | M18 | 25 | 4 | 96 | “ | |||||
| “ | Italy | M19 | 40 | 15 | 85 | “ | |||||
| “ | Sardinia | M20 | 96 | 10 | 90 | “ | |||||
| “ | Sardinia | M21 | 68 | 9 | 91 | “ | |||||
| “ | Iceland | S1 | 30 | 100 | “ | ||||||
| “ | Denmark | S2-3-42 | 105 | 100 | “ | ||||||
| “ | Netherlands | S5-6r-7 | 162 | 100 | “ | ||||||
| “ | Germany | S8 | 85 | 100 | |||||||
| “ | Switzerland | S9-10-11 | 167 | 100 | “ | ||||||
| “ | Italy | A1-2-3-4-5 | 130 | 100 | “ | ||||||
| “ | Italy | A6-7-8-9 | 253 | 100 | “ | ||||||
| “ | Sicily | A10-11 | 83 | 100 | “ | ||||||
| Total | 4416 | 45 | 53 | 0.27 | 0.09 | 0.07 | 0.36 | 1.8 | |||
| . | . | . | . | Percentage . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Location . | n . | XY . | XX . | XO . | OY . | XXX . | XXY . | YY . | Reference . | ||
| Africa | S. Africa | SA1 | 31 | 90 | 10 | (Denholm et al. 1990) | |||||
| “ | “ | SA2 | 33 | 30 | 64 | 6 | “ | ||||
| Asia/Europe | Turkey | Antalya | 30 | 53 | 47 | (Cakir and Kence 1996) | |||||
| “ | “ | Incekum | 31 | 74 | 26 | “ | |||||
| “ | “ | Anamur | 31 | 77 | 23 | “ | |||||
| “ | “ | Gulnar | 32 | 34 | 66 | “ | |||||
| “ | “ | Kayrak | 30 | 10 | 90 | “ | |||||
| “ | “ | Y. Cadiri | 32 | 41 | 59 | “ | |||||
| “ | “ | Silifke | 30 | 30 | 70 | “ | |||||
| “ | “ | Atakent | 30 | 60 | 40 | “ | |||||
| “ | “ | Mersin | 36 | 42 | 58 | “ | |||||
| “ | “ | Adana | 30 | 43 | 57 | “ | |||||
| “ | “ | Yumurtalik | 30 | 23 | 77 | “ | |||||
| “ | “ | Karatas | 25 | 56 | 44 | “ | |||||
| “ | “ | Ceyhan | 30 | 70 | 30 | “ | |||||
| “ | “ | Samsun | 32 | 75 | 25 | “ | |||||
| “ | “ | Giresun | 30 | 0 | 100 | “ | |||||
| “ | “ | Trabzon | 33 | 0 | 100 | “ | |||||
| “ | “ | Rize | 30 | 13 | 87 | “ | |||||
| “ | “ | Artvin | 33 | 24 | 76 | “ | |||||
| “ | “ | Erzurum | 32 | 53 | 47 | “ | |||||
| “ | “ | Erzincan | 36 | 83 | 17 | “ | |||||
| “ | “ | Sivas | 33 | 91 | 9 | “ | |||||
| “ | “ | Yozgat | 30 | 77 | 23 | “ | |||||
| “ | “ | Izmit | 30 | 100 | “ | ||||||
| “ | “ | Isparta | 30 | 40 | 60 | “ | |||||
| “ | “ | Bursa | 31 | 68 | 32 | “ | |||||
| “ | “ | Tokat | 36 | 69 | 31 | “ | |||||
| “ | “ | Istanbul | 30 | 23 | 77 | “ | |||||
| “ | “ | Iskenderun | 29 | 24 | 76 | “ | |||||
| “ | “ | Afyon | 28 | 61 | 39 | “ | |||||
| “ | “ | Usak | 32 | 50 | 50 | “ | |||||
| “ | “ | Ismir | 31 | 29 | 71 | “ | |||||
| “ | “ | Manisa | 29 | 55 | 45 | “ | |||||
| “ | “ | Balikesir | 28 | 25 | 75 | “ | |||||
| “ | “ | Simav | 29 | 30 | 70 | “ | |||||
| “ | “ | Ankara | 31 | 71 | 29 | “ | |||||
| “ | “ | Polatli | 31 | 97 | 3 | “ | |||||
| Europe | UK | Fm 42 | 51 | 88 | 4 | 8 | (Denholm et al. 1985) | ||||
| “ | “ | Fm 39 | 33 | 58 | 39 | 3 | “ | ||||
| “ | “ | Fm 31 | 28 | 21 | 79 | “ | |||||
| “ | “ | Fm 44 | 47 | 98 | 2 | “ | |||||
| “ | “ | Fm 45 | 48 | 75 | 19 | 4 | 2 | “ | |||
| “ | “ | Harpenden | 223 | 5 | 93 | 2 | 0 | “ | |||
| “ | “ | Fm 3 | 19 | 21 | 79 | (Denholm et al. 1983) | |||||
| “ | “ | Fm 9 | 33 | 100 | “ | ||||||
| “ | “ | Fm 6 | 36 | 89 | 11 | “ | |||||
| “ | “ | Fm 13 | 29 | 100 | “ | ||||||
| “ | “ | Fm 11 | 11 | 100 | “ | ||||||
| “ | “ | Fm 14 | 27 | 11 | 89 | “ | |||||
| “ | “ | Fm 22 | 46 | 2 | 98 | “ | |||||
| “ | “ | Fm 29 | 22 | 14 | 82 | 5 | “ | ||||
| “ | France | M1 | 87 | 77 | 15 | 8 | (Franco et al. 1982) | ||||
| “ | France | M2 | 49 | 59 | 41 | “ | |||||
| “ | Yugoslavia | M3 | 69 | 46 | 52 | 1 | “ | ||||
| “ | Italy | M4 | 92 | 85 | 15 | “ | |||||
| “ | Italy | M5 (2r) | 178 | 88 | 12 | “ | |||||
| “ | Italy | M5 (2r) | 94 | 67 | 17 | 16 | “ | ||||
| “ | Italy | M6 | 44 | 50 | 50 | “ | |||||
| “ | Italy | M7 (2r) | 149 | 64 | 36 | 1 | “ | ||||
| “ | Italy | M8 | 56 | 84 | 7 | 9 | “ | ||||
| “ | Italy | M9 (2) | 52 | 83 | 15 | 2 | “ | ||||
| “ | Italy | M10 | 72 | 31 | 67 | 3 | “ | ||||
| “ | Italy | M11 | 46 | 4 | 96 | “ | |||||
| “ | Italy | M12 | 61 | 39 | 57 | 3 | “ | ||||
| “ | Italy | M13 | 63 | 2 | 92 | 6 | “ | ||||
| “ | Italy | M14 | 72 | 19 | 81 | “ | |||||
| “ | Italy | M15 | 43 | 56 | 44 | “ | |||||
| “ | Italy | M16 | 54 | 35 | 61 | 4 | “ | ||||
| “ | Italy | M17 | 62 | 2 | 98 | “ | |||||
| “ | Italy | M18 | 25 | 4 | 96 | “ | |||||
| “ | Italy | M19 | 40 | 15 | 85 | “ | |||||
| “ | Sardinia | M20 | 96 | 10 | 90 | “ | |||||
| “ | Sardinia | M21 | 68 | 9 | 91 | “ | |||||
| “ | Iceland | S1 | 30 | 100 | “ | ||||||
| “ | Denmark | S2-3-42 | 105 | 100 | “ | ||||||
| “ | Netherlands | S5-6r-7 | 162 | 100 | “ | ||||||
| “ | Germany | S8 | 85 | 100 | |||||||
| “ | Switzerland | S9-10-11 | 167 | 100 | “ | ||||||
| “ | Italy | A1-2-3-4-5 | 130 | 100 | “ | ||||||
| “ | Italy | A6-7-8-9 | 253 | 100 | “ | ||||||
| “ | Sicily | A10-11 | 83 | 100 | “ | ||||||
| Total | 4416 | 45 | 53 | 0.27 | 0.09 | 0.07 | 0.36 | 1.8 | |||
Blank cells equal 0%.
In addition to the variation in M and Md-traD observed in natural populations, other alleles of both genes have been isolated in the laboratory. A loss-of-function mutation in Md-tra, Md-traman (formerly Fman), turns wild-type Md-tra into a female-determining allele in the absence of M (Schmidt et al. 1997a). The Md-traman mutation removes TRA/TRA2 binding sites, which likely prevents TRA from autoregulating the splicing of the Md-traman allele into a functional female-specific isoform (Hediger et al. 2010). In laboratory strains carrying the Md-traman mutation and lacking any M factors, males were the homogametic sex (homozygous for Md-traman) and females were heterogametic (Md-traman/Md-tra+) (Schmidt et al. 1997a).
The activity of Md-tra can be inhibited by mutations that affect the maternal germline or zygote separately, which can convert the house fly sex determination system into one controlled by the maternal genotype. A recessive mutation on chromosome IV, which is likely a hypomorphic allele of Md-tra that is lacking maternal germline function (Schmidt et al. 1997a), was fortuitously named transformer (tra) by Inoue and Hiroyoshi (1986). Homozygous females (tra/tra) produced intersexes or males without an M factor, whereas heterozygotes (tra/+) produced mostly females when mated to males lacking M (Inoue and Hiroyoshi 1986). Zygotes carrying an M factor developed into males regardless of whether the mother had one or two copies of this tra mutation (Inoue and Hiroyoshi 1986). The house fly tra mutation produces a situation reminiscent of that observed in naturally occurring systems in which maternal genotype determines sex, such as Chrysomya rufifacies, suggesting a mutational mechanism by which maternal effect sex determination can evolve (Scott et al. 2014b).
Similarly, the germline and zygotic activity of M can be separated, also creating house fly strains with maternal effect sex determination. The Arrhenogenic (Ag) mutation (Vanossi and Rovati 1982) on the first chromosome was most likely a hypomorphic allele in the promoter region of an M locus on chromosome I that maintained germline expression but impaired expression in the zygote (Schmidt et al. 1997a; Dübendorfer et al. 2002). Females that were heterozygous for the Ag allele failed to activate Md-tra in the germline (because the germline activity of M inhibits Md-tra) and produced all male offspring (Hediger et al. 2010). When this strain was maintained in the laboratory, homozygous wild-type females produced all-female offspring because males of this strain lack an M allele with zygotic activity (Hediger et al. 2004).
Early studies revealed a virtual lack of crossing-over in male house flies (McDonald 1971; Lester et al. 1979), consistent with what is observed in most other dipterans (White 1973; Gethmann 1988). This facilitated genetic studies to determine the chromosomal locations of sex-determining factors. Later worked revealed that the crossover frequencies in males vary, depending on the genes examined and the populations used. Reported values range between 0–0.53% (Hamm et al. 2005; Hamm 2008), 0.03–0.11% (Sullivan 1961), 9.3–31% (Lester et al. 1979), and 7–28% (Feldmeyer et al. 2010). Intriguingly, greater male recombination rates tend to be associated with AM (Sullivan 1961; Hiroyoshi et al. 1982; Inoue and Hiroyoshi 1982; Gethmann 1988; Hamm et al. 2005; Hamm 2008; Feldmeyer et al. 2010).
Evidence for male recombination in laboratory experiments could be the result of meiotic crossing over and/or premeiotic events such as mitotic recombination. Genomic rearrangements, Y-autosome translocations, mobile element insertions, and transposable M factors increase the frequency of male recombination in multiple different dipteran species, but this is not necessarily because of an increased rate of meiotic recombination (Gethmann 1988). Asymmetrical reciprocal recombinant classes suggest that many examples of male recombination in house fly might be the result of premeiotic events (e.g., mitotic recombination) or aneuploid segregants, not meiotic recombination (Rubini et al. 1980; Gethmann 1988). Megaselia scalaris also has a transposing male-determining factor, and male recombination in this species appears to result from premeiotic events (Gethmann 1988). A nonrandom association between transposition of the M. scalaris M and male recombination suggest that the two processes may be caused by similar underlying factors in the male germline (Mainx 1964). This parallels the association between transposable element derepression and male recombination observed in Drosophila, suggesting a common effect of transposable elements and transposing M factors on genome instability in the premeiotic male germline (Gethmann 1988). Alternatively, elevated male recombination in AM genotypes might reflect the early stages of differentiation in a nascent sex chromosome system where male recombination is not yet repressed (Feldmeyer et al. 2010). Additional experiments are needed to test these hypotheses.
Geographic distribution of AMvs. YM and/or XM males
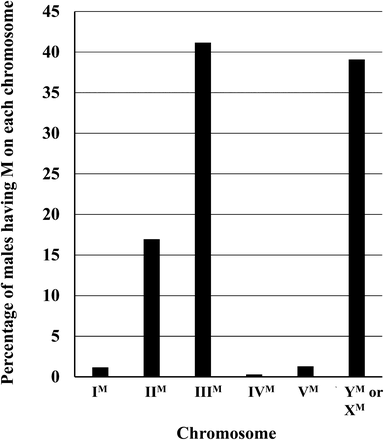
Non-YM house fly populations exist in nature throughout the world, and M factors can be found at a wide range of frequencies. A summary of the papers reporting on the linkage of M is given in Table 1. AM males have been found on most continents, with the notable exception of Central and South America, for which there have been no published studies. M has been found most frequently on autosome III, followed by the Y or X chromosome, and then autosome II (Figure 3). M is rarely found on autosomes I, IV, or V (Figure 3). Most studies that do not find M on an autosome assume that this is a YM strain, based on the belief this is the ancestral condition. However, M may also be X-linked, so without karyotyping, distinguishing between YM and XM is not possible. A summary of the different karyotypes found for male house flies (Table 2) reveals the frequencies of YM and XM are about equal across the populations surveyed. Other male karyotypes (e.g., XO, OY, XXX, XXY, YY) were detected (Table 2), but were overall rare (<2%). There are also many populations that contain males with multiple M factors. The Ipswich (Australia) population has the highest number of multiple M males and homozygous M males found to date—92 and 70%, respectively (Hamm and Scott 2009). This represents an extreme case and one in which the population appears headed for females to become the heterogametic sex. The presence of Md-traD was confirmed in this population, although the frequency of Md-traD was not determined (Hamm and Scott 2009).
The relative percentage of males with M on each of the chromosomes. Results were calculated from the data in Table 1. Values represent relative percentages, as different reports used in Table 1 accounted for males with multiple M factors using different calculations. Studies failing to find a linkage of M to an autosome called these strains YM, although in the absence of karyotype information these strains could also be XM.
Like AM males, females with Md-traD have also been found throughout the world (Table 1). The frequency of Md-traD in females varies from 0% in some populations to 100% of the females in four locations in Tanzania. It would be expected that all males would either carry multiple M factors or be homozygous for M in populations where Md-traD is found in all females, but unfortunately this was not investigated in these studies. One Md-traD haplotype (accession# GU070694) contains three small intronic insertions/deletions (indels), a small insertion in a male-specific exon, and one nonsynonymous substitution in the coding region (Hediger et al. 2010). The indels are thought to allow for the zygotic splicing of the Md-traD allele into a functional isoform in the absence of the feed-forward activity of Md-tra from the maternal germline, and they may prevent the negative regulation of M (Hediger et al. 2010) (Figure 1E). The same Md-traD haplotype was found in seven different populations sampled across Europe, North America, Asia, Africa, and Australia (Scott et al. 2014a). In contrast, multiple Md-tra+ haplotypes were found in these populations, leading to speculation that Md-traD may have a single evolutionary origin followed by a recent global spread.
Surveys of house flies on multiple continents (in the northern hemisphere) have revealed latitudinal gradients of AM and YM populations, with YM males most common in the north and AM or XM more common in the south. For example, in European populations ranging from Sicily to Denmark and Iceland, Franco et al. (1982) found AM and XM males more often below the 44th parallel and YM males more frequent in the north. McDonald et al. (1975) reported latitudinal variation in AM in North America, with populations from North Dakota, Texas, and Florida containing IIIM males at 0.8%, 10.4%, and 100%, respectively. However, this study did not survey M factors on other autosomes and found a low frequency of AM males in Texas, a southern location. Stronger evidence for a North American latitudinal gradient in AM was revealed in a survey that sampled from Florida (29° 41′ latitude) to Maine (44° 2′) (Hamm et al. 2005). In Florida, 100% of the males possess the M factor on chromosome III. North Carolina had 20% IIIM and 2.35% with both YM and IIIM in the same individual. Fewer IIIM males were located in New York (4.35%), and the Maine population was entirely XYM or XXM. This range in latitude was similar to that in Japan, where a north-south cline was also observed (Tomita and Wada 1989b). Md-traD is distributed sporadically throughout the Japanese populations at frequencies ranging between 0 and 99% of females (Tomita and Wada 1989b).
Other patterns have been observed in the spatial distribution of AM. For example, a radial cline was detected in the British Isles (Denholm et al. 1985). Populations in central England were predominantly XMXM, whereas XYM males were found to inhabit the north. The frequency of Md-traD, XM, and a rarer IIIM decreased on moving north, east, and west. Karyotype data revealed YM to be extremely rare in most strains collected in the south of England, increasing in frequency upon moving north. The Y chromosome morphology appeared small in southeast England, and the longest Y chromosome was observed in Scotland (the north). Two sites at the same latitude differed in the frequency of the Y chromosome, supporting the radial cline hypothesis.
Franco et al. (1982) also reported an altitudinal gradient in Europe, with AM populations less than 100 meters above sea level. AM males decreased as the altitude increased. An altitudinal cline also was detected in Turkey. Cytological examinations revealed frequencies of XX males (assumed to be AM) ranging from 3.22 to 100% (Cakir and Kence 1996), and XX males were present in 10 of the 36 populations at frequencies greater than 70% (Table 2). There were fewer XX males in the central and eastern Anatolian highlands than in the coastal regions (Cakir and Kence 1996). The Y chromosome was absent in three populations (Izmit, Giresun, and Trabzon). Further research in Turkey established the existence of IIIM, VM, and Md-traD (Cakir 1999) (Table 1).
Strains with different numbers of M factors have the ability to produce a variety of sex ratios depending on their genetic makeup (Table 3). According to Fisher’s theory, the equilibrium sex ratio is most likely to be 1:1 due to the notion that if one sex is rare, it will have greater reproductive success (Goodenough et al. 1993), and a modeling study in house fly supported this optimal ratio for house flies (Kozielska et al. 2006). The most common way to maintain equal sex ratios is for parents to have equal numbers of male and female offspring, and any deviation should be automatically corrected by selection in favor of the other sex (Fisher 1930; Hamilton 1967). In house flies, populations that contain only one M factor found only in a heterozygous state (either Y-linked, X-linked, or autosomal) will produce a 1:1 ratio of males to females. However, deviations from a 1:1 sex ratio can be obtained when a male carries multiple M factors (Table 3). If a normal female produces only sons, her mate must be homozygous for at least one AM (or XM). This male may or may not have additional M factors. A male heterozygous for the M factor on two different chromosomes will produce 75% male offspring, whereas a male with five M factors in heterozygous form will produce 96.9% male offspring (Table 3). These situations all assume that the female does not carry Md-traD. The house fly sex determination polymorphism therefore provides a mechanism by which biased sex ratios are produced in the absence of meiotic drive or some other non-Mendelian sex-ratio distortion system.
Examples of the different percentages of males produced by different male genotypes assuming that the population lacks Md-traD
| Male Genotype . | % males in F1 . |
|---|---|
| IIIM/III | 50 |
| XYM | 50 |
| IIM/II;IIIM/III | 75 |
| IIIM/III;XYM | 75 |
| IIM/II;IIIM/III;/IVM/IV | 87.5 |
| IIIM/III;IVM/IV;XYM | 87.5 |
| IM/I;IIM/II;IIIM/III;IVM/IV | 93.4 |
| IM/I;IIM/II;IIIM/III;IVM/IV;VM/V | 96.9 |
| IIM/II;IIIM/IIIM;XYM | 100 |
| IIIM/IIIM | 100 |
| IIM/II;IIIM/IIIM | 100 |
| IIM/IIM;IIIM/IIIM | 100 |
| IIM/II;IIIM/IIIM;IVM/IV;XYM | 100 |
| Male Genotype . | % males in F1 . |
|---|---|
| IIIM/III | 50 |
| XYM | 50 |
| IIM/II;IIIM/III | 75 |
| IIIM/III;XYM | 75 |
| IIM/II;IIIM/III;/IVM/IV | 87.5 |
| IIIM/III;IVM/IV;XYM | 87.5 |
| IM/I;IIM/II;IIIM/III;IVM/IV | 93.4 |
| IM/I;IIM/II;IIIM/III;IVM/IV;VM/V | 96.9 |
| IIM/II;IIIM/IIIM;XYM | 100 |
| IIIM/IIIM | 100 |
| IIM/II;IIIM/IIIM | 100 |
| IIM/IIM;IIIM/IIIM | 100 |
| IIM/II;IIIM/IIIM;IVM/IV;XYM | 100 |
Nearly all of these genotypes have been observed in field collected flies, although others exist as well (Hamm et al. 2005; Hamm and Scott 2008, 2009). In theory, any of the five autosomes could exhibit these genotypes and produce the same proportion of male offspring (e.g., IIIM/IIIM or VM/VM both produce only male progeny in the absence of Md-traD).
| Male Genotype . | % males in F1 . |
|---|---|
| IIIM/III | 50 |
| XYM | 50 |
| IIM/II;IIIM/III | 75 |
| IIIM/III;XYM | 75 |
| IIM/II;IIIM/III;/IVM/IV | 87.5 |
| IIIM/III;IVM/IV;XYM | 87.5 |
| IM/I;IIM/II;IIIM/III;IVM/IV | 93.4 |
| IM/I;IIM/II;IIIM/III;IVM/IV;VM/V | 96.9 |
| IIM/II;IIIM/IIIM;XYM | 100 |
| IIIM/IIIM | 100 |
| IIM/II;IIIM/IIIM | 100 |
| IIM/IIM;IIIM/IIIM | 100 |
| IIM/II;IIIM/IIIM;IVM/IV;XYM | 100 |
| Male Genotype . | % males in F1 . |
|---|---|
| IIIM/III | 50 |
| XYM | 50 |
| IIM/II;IIIM/III | 75 |
| IIIM/III;XYM | 75 |
| IIM/II;IIIM/III;/IVM/IV | 87.5 |
| IIIM/III;IVM/IV;XYM | 87.5 |
| IM/I;IIM/II;IIIM/III;IVM/IV | 93.4 |
| IM/I;IIM/II;IIIM/III;IVM/IV;VM/V | 96.9 |
| IIM/II;IIIM/IIIM;XYM | 100 |
| IIIM/IIIM | 100 |
| IIM/II;IIIM/IIIM | 100 |
| IIM/IIM;IIIM/IIIM | 100 |
| IIM/II;IIIM/IIIM;IVM/IV;XYM | 100 |
Nearly all of these genotypes have been observed in field collected flies, although others exist as well (Hamm et al. 2005; Hamm and Scott 2008, 2009). In theory, any of the five autosomes could exhibit these genotypes and produce the same proportion of male offspring (e.g., IIIM/IIIM or VM/VM both produce only male progeny in the absence of Md-traD).
Little evidence for changes in frequency of AM males over time in field populations
Despite the variation that occurs between populations, studies on the relative frequency of AMvs. YM over time within field populations, from the United States and Europe, have shown that the populations are relatively unchanged from the 1970s onwards. Male flies collected in 1973, 2003, and 2009 from Florida were 100% IIIM (McDonald et al. 1975; Hamm et al. 2005; Kavi et al. 2014). The frequency of AM males in Europe was evaluated in 2006 by the use of 15 collections from southern Italy to northern Germany and compared with collections made 25 years earlier. There was no clear change in the distribution of sex-determining factors (Kozielska et al. 2008). In flies from North Carolina, frequencies of III/III; XYM, IIIM/III; XX, and IIIM/III; XYM males were unchanged (karyotypes were assumed, but not determined) between 2002 and 2006 (Hamm and Scott 2008). Field-collected flies from this population in 2007 showed a slight increase in the frequency of XYM males and a slight decrease in the frequency of IIIM/III males (relative to 2002 and 2006), suggesting that the relative frequency of XYM and IIIM/III can vary slightly over time (Hamm and Scott 2008). The first recorded autosomal male (IIIM) factor in the northeast United States was reported in 2003 (Shono and Scott 2003) from flies collected in New York (and laboratory selected with the insecticide spinosad). In contrast, field-collected flies from New York in 1980 (Scott et al. 1984) and 1987 (Konno and Scott 1991) (that were also selected with insecticides) were XYM or XXM, leading to the suggestion that the frequency of AM might be increasing (Shono and Scott 2003). A 2005 study showed that flies from New York were IIIM at a frequency of 4.35% of the population (Hamm et al. 2005), so it is unclear whether the failure to detect AM males in 1980 and 1987 was due to the low frequency of AM or if the frequency is actually increasing.
In addition to field-collected strains, the linkage of M has been determined in several laboratory strains. These results are summarized in Supporting Information, Table S1. These data, particularly if the collection site is known, can provide additional information about the distribution of AM males. However, colonization in the laboratory will likely alter the frequency of the different M factors (Hamm and Scott 2008). The frequency of the linkage of M in laboratory strains was similar to that found for field collections, with IIIM and YM being the most common. Curiously, M in the SRS strain maintained by different laboratories has been linked to V (Hamm et al. 2005), Y (Milani et al. 1967; Franco et al. 1982), and III (Hamm 2008). It is difficult to assess whether these differences are attributable to local adaptation or separate contamination events.
Studies of the relative fitness of AM and YM males
Franco et al. (1982) noted that “all the papers concerning the karyotype of Musca domestica L. (2n = 12) published between 1908 and 1948 . . . reported the presence of XX females and XY males . . . It can be assumed that the authors, being European, examined houseflies of European origin. Since 1958, cases of sex-limited inheritance, interpreted a posteriori as due to autosomal sex-determinants, have been described in several strains of houseflies of non-European origin.” Starting in about 1960, the reports of AM males increased, but it is not clear whether this was a result of the recent invasion of autosomal M factors, incomplete sampling in earlier studies, or neglecting to search for AM. This spread of AM males (perceived or real) led to the suggestion that it might be causally related to selection for insecticide resistance (Hiroyoshi 1980), although later the author no longer held that opinion (personal communication to R. M. Sawicki, cited in Denholm et al. 1983).
Insecticide resistance in the house fly has been studied widely and is most commonly not sex-linked (Tsukamoto 1983), although there are some exceptions. One study found that the frequency of IIIM males increased after selection with insecticide (permethrin), and the authors concluded this could be due to either tight linkage between the locus conferring resistance and the IIIM locus or to genetic drift (Denholm et al. 1983). A study directly comparing insecticide resistance levels and frequency of AM males in four geographically separate populations found no correlation between resistance (including kdr-type resistance on chromosome III) and the frequency of AM (or IIIM) males (Hamm et al. 2005). Although there is an important mechanism of pyrethroid resistance on autosome III (kdr-type), this resistance is inherited as an incompletely recessive trait (i.e., heterozygotes have only low levels of resistance) (Shono 1985). It is therefore unlikely that selection for pyrethroid resistance in heterozygotes drove the invasion of IIIM. However, there are two reports of sex-linked (male-limited) inheritance of insecticide resistance in natural populations (Kerr 1960; Kence and Kence 1992). The first was a report of about eightfold greater resistance to dichlorodiphenyltrichloroethane in males than females in the Canberra strain, but the linkage of resistance and M was not reported (Kerr 1960). The second was a report of greater levels of malathion resistance in males than females in F1 male backcross progeny of the resistant Ankara strain and a susceptible marker strain. The resistance was linked to autosomes II and V, and the strain was IIM (Kence and Kence 1992). These authors suggested that the linkage of M in the Ankara strain had shifted from IIIM to IIM as a result of the malathion selection. It is therefore conceivable that an autosomal M factor could invade a natural population through linkage with an insecticide resistance allele, but selection for insecticide resistance cannot explain most of the autosomal M polymorphisms.
The geographical variation in the distribution of AM and YM suggests that selection may be acting on fitness differences associated with different M genotypes in different environments. Fitness can be used to describe a variety of characteristics including, but not limited to, fecundity, emergence time, mating success, size, longevity, or susceptibility to disease. Deviations from random mating can be attributed to a difference in female receptivity, preferential mating within strains, or in male competition. There are important aspects of house fly biology that pertain to fitness of YM, XM, and AM males. House flies can survive the winter in cold climates as small populations living indoors, especially at livestock facilities (Keiding 1986). Black and Krafsur (1986) looked at seasonal house fly reproduction at one dairy and three swine farrowing sheds. They found slowed reproduction at the dairy in winter and early spring due to chronically low temperatures. House flies cannot survive freezing temperatures and do not diapause (Black and Krafsur 1986; Keiding 1986). House fly overwintering sites must offer microhabitats that remain greater than −5° with sufficient time greater than 10° (Rosales et al. 1994). Adults will mate within the first day after eclosion if adequate food is available (Milani 1975). The average mating speed for single pair crosses was found to be about 30 min and copulation lasts more than 1 hr (Bryant 1980). Females will only mate once unless additional sperm are necessary for further egg production (Keiding 1986). A recopulation frequency of 3.7% was determined (Baldwin and Bryant 1981). Genotype by environment fitness effects associated with any of these aspects of house fly biology could be responsible for the invasion of AM and/or the maintenance of spatial gradients.
The relative fitness of YMvs. IIIM males has been compared with the use of isogenic strains that carried the IIIM or YM chromosome (Hamm et al. 2009). Three different comparisons were made. First, cages were started with 50% YM and 50% IIIM males, and the frequencies of YM and IIIM males were evaluated across generations. Second, mating competition studies were performed. Third, the relative emergence rates of IIIMvs. YM pupae were examined at four temperatures. All three studies found that IIIM males had a greater fitness than YM males. In the cage competition studies, >90% of the males were IIIM after seven generations. IIIM males were more likely to mate than YM males, and a greater percent of IIIM males emerged after being held as pupae at 4, 16, or 28° for 3 d (Hamm et al. 2009).
In contrast to the aforementioned experiments, a comparison of the frequency of AM and YM males in houseflies after 4 yr in the laboratory found a selective disadvantage for IIIM males (Hamm and Scott 2008). In 2002, 77.7% of the male house flies were III/III;XYM, 20% were IIIM/III;XX, and 2.3% were IIIM/III;XYM (karyotypes were inferred, not determined). After 4 yr in the laboratory, IIIM/III males disappeared and all of the males were either XYM (82.6%) or XMYM (17.4%). There are at least four possible explanations why there was strong selection against IIIM males in this laboratory experiment (Hamm and Scott 2008), but selection in favor of the IIIM chromosome in the studies using the isogenic strains (Hamm et al. 2009): 1) the field collected strain that was left in the laboratory for 4 yr (Hamm and Scott 2008) contained Md-traD; 2) there were four male genotypes in the 2008 study, but only two in the 2009 study (thus, the competition was not exactly the same); 3) the two papers used strains with different genetic backgrounds, which could influence the relative fitness; and 4) the 2008 study, as a whole, was not replicated.
The availability of the house fly genome sequence (Scott et al. 2014a) will open up new avenues of experimentation to further pursue fitness differences between YM and AM males. For example, there is evidence for gene expression differences between YM and IIIM males, which could be responsible for phenotypic differences that may be under selection (R. P. Meisel, J. G. Scott, and A. G. Clark, unpublished data).
Why are there AM and YM populations?
Ever since the discovery of differences between populations in the frequencies of AM and YM males, researchers have struggled to understand the forces responsible for the patterns observed. Understanding the factors responsible for the invasion of new male- and female-determining loci in house fly and the maintenance of polygenic sex determination could reveal generalizable insights into the factors responsible for the evolution of sex determination.
The north-south clines (AM in the south and YM in the north) observed in the Northern hemisphere (Franco et al. 1982; Tomita and Wada 1989b; Hamm et al. 2005) and the southern hemisphere (YM in the south and AM in the north) (Feldmeyer et al. 2008) are best explained by seasonality in temperature variation, whereas variation in Md-traD is best explained by variation in humidity and yearly mean temperature (Feldmeyer et al. 2008). This suggests that autosomal M factors may be linked to allelic variation with ecologically adaptive fitness effects. Other types of clines also have been observed (e.g., radial), which suggests additional environmental variables may be associated with the distribution of AM and YM. Although it also was hypothesized that increases in AM could be correlated with insecticide resistance (Hiroyoshi 1980; Franco et al. 1982; Kence and Kence 1992), this does not appear to be the case for reasons discussed above.
The theoretical model of Bull and Charnov (1977) suggests two stages for the transition between standard populations (XYM males and XX females) and populations fixed for an autosomal male determining locus (AM). In the first stage, an invading autosomal male determiner either confers a fitness benefit or is genetically linked to a beneficial allele, and it increases in frequency. In the second stage, an epistatic female determining factor (e.g., Md-traD) invades and allows for the fixation of the autosomal male determiner.
Bull and Charnov (1977) only modeled the invasion of new sex determining loci via natural selection, and they did not consider the role of sex ratio selection in the invasion of the female determining locus. However, subsequent work demonstrated that sex ratio selection could not cause a complete transition between sex determination systems in house fly, but it can affect the frequency of sex determining loci in populations (Kozielska et al. 2006). In addition, the Bull and Charnov (1977) model predicts equilibria in which polygenic sex determination is maintained (i.e., the AM locus does not fix). The altitudinal, latitudinal, and radial variation in AM frequencies could be interpreted as either populations at a polygenic equilibrium or transient states on the way to fixation of AM. If these populations are on the way to fixation of AM, the relative stability of populations over generations suggests that this process is moving slowly.
Unanswered questions and future directions
The complexity of sex determination in the house fly has left several unanswered questions. Many areas have not been considered or tested. Are autosomal M factors moving through populations because of a selective advantage? If a selective advantage is present, what phenotypes are under selection? Is selection acting directly on the phenotypic effects of different M and Md-tra alleles/loci, or does selection act on allelic variants genetically linked to the M or Md-tra loci? It is important to determine what the selection pressures are and how they vary in different environments, leading to populations that have varied frequencies of Md-traD, M factors and linkage of M. These results would allow us to test models for the evolution of sex determination, providing novel insights into the factors responsible for the evolution of sex determination pathways.
It is surprising that the linkage of M and frequency of females with Md-traD has not been investigated in Central or South America. This is a gap in our knowledge that would be useful to fill because it would provide an additional independent test of geographic clines in the frequency of AM. In addition, there are relatively few studies that have determined the frequency of males with multiple M factors and/or the frequency of females with Md-traD. More studies of this type will help understand how these genes co-evolve.
What is M? Identification of the M factor would be a tremendous advance for understanding house fly sex determination and the nature of sex determination pathways in general. Is the M factor a mobile element that can transpose between chromosomes or is each instance of M on a different chromosome a unique gene that has gained the ability to negatively regulate Md-tra? Is the same gene used in other dipteran sex determination pathways as a male determining locus? Knowing the identity of M would allow us to test whether the variability in the frequency of M on different chromosomes is a result of fitness effects of different alleles of M on each chromosome or selection on allelic variation in genes linked to the autosomal M loci.
Although several studies have found populations in which M is not linked to an autosome, clarification as to whether such populations are YM or XM would be helpful. If we knew the sequence of M, we also could potentially identify chromosome-specific allelic variants of M. That would allow us to diagnose the location of M through genotyping by sequencing (potentially being able to diagnose the locations of multiple M factors in an individual as well). We could then sample old specimens (e.g., from insect collections) to shed light on the relative frequency of M on different chromosomes in populations from the 1800s and 1900s. Efforts to karyotype house flies are laborious and substantial time must be spent learning how to correctly assess the patterns of chromosomes in the squashes. Having visible labels, stains, or molecular markers for specific chromosomes, especially X and Y, would greatly facilitate obtaining the proper karyotype (and would move this area of investigation forward at a more rapid pace).
The genome sequence of the house fly will allow for investigations that test for early differentiation of nascent sex chromosomes. Studying such “neo-X” and “neo-Y” chromosomes has been a fruitful area of research in Drosophila genetics to characterize the evolutionary forces that act upon X and Y chromosomes (Sturgill et al. 2007; Meisel et al. 2009; Zhou and Bachtrog 2012). There are many theoretical predictions about how mutation, selection, recombination, and genetic drift drive the differentiation of sex chromosomes (Vicoso and Charlesworth 2006), and the house fly is poised to be a unique model for investigating the early stages of this important evolutionary process.
The house fly is a serious threat to human and animal health. Adult house flies are vectors of more than 100 human and animal intestinal diseases (Scott and Lettig 1962; Greenberg 1965; Keiding 1986). They are capable of transmitting parasites that cause typhoid fever, cholera, bacillary dysentery, infantile diarrhea, tuberculosis, plague, leprosy, yaws, salmonellosis, anthrax, and other diseases (West 1951). Flies also transmit eye diseases such as trachoma and epidemic conjunctivitis (Keiding 1986). Therefore, control of house flies is an area of great significance, but most approaches rely on the use of insecticides which present environmental and health concerns. Release of sterile males has been a great success for some Diptera, such as screw worm (Cochliomyia hominivorax) (Knipling 1960). An understanding of the factors underlying the relative frequency of YM and AM, as well as the identification of M may offer new insights into fly reproduction that could lead to new control methods, such as the release of homozygous sterile M males into closed systems, such as poultry facilities. This would lead the following generation to produce all males, providing control of the population. Additional strategies will follow as a deeper understanding of this biological system is attained.
Acknowledgments
This work was supported by Hatch Project NYC-139416 to J.G.S.
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.014795/-/DC1
Communicating editor: J. M. Comeron
Literature Cited
Hamm, R. L., 2008 Exploring the population genetics of the house fly sex determining genes, M and F. Ph.D. Thesis, Cornell University, Ithaca, NY.
Hiroyoshi, T., 1980 Formal genetics of the housefly in relation to insecticide resistance, pp. 397 in Proceedings of the 16th International Congress of Entomology, Elsevier Biomedical, Kyoto, Japan.